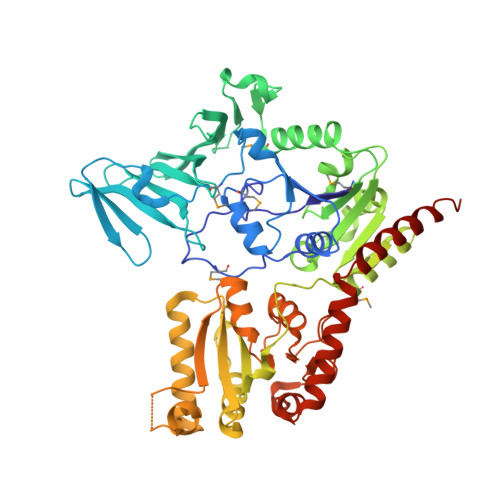
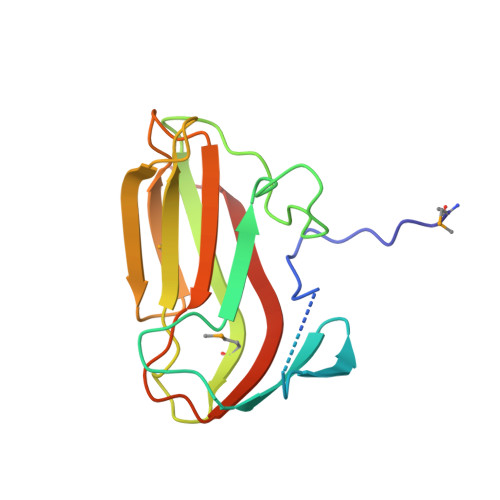
Unraveling the sequence of cytosolic reactions in the export of GspB adhesin fromStreptococcus gordonii.
Chen, Y., Bensing, B.A., Seepersaud, R., Mi, W., Liao, M., Jeffrey, P.D., Shajahan, A., Sonon, R.N., Azadi, P., Sullam, P.M., Rapoport, T.A.(2018) J Biol Chem 293: 5360-5373
- PubMed: 29462788
- DOI: https://doi.org/10.1074/jbc.RA117.000963
- Primary Citation of Related Structures:
5VAE, 5VAF - PubMed Abstract:
Many pathogenic bacteria, including Streptococcus gordonii , possess a pathway for the cellular export of a single serine-rich-repeat protein that mediates the adhesion of bacteria to host cells and the extracellular matrix. This adhesin protein is O -glycosylated by several cytosolic glycosyltransferases and requires three accessory Sec proteins (Asp1-3) for export, but how the adhesin protein is processed for export is not well understood. Here, we report that the S. gordonii adhesin GspB is sequentially O -glycosylated by three enzymes (GtfA/B, Nss, and Gly) that attach N -acetylglucosamine and glucose to Ser/Thr residues. We also found that modified GspB is transferred from the last glycosyltransferase to the Asp1/2/3 complex. Crystal structures revealed that both Asp1 and Asp3 are related to carbohydrate-binding proteins, suggesting that they interact with carbohydrates and bind glycosylated adhesin, a notion that was supported by further analyses. We further observed that Asp1 also has an affinity for phospholipids, which is attenuated by Asp2. In summary, our findings support a model in which the GspB adhesin is sequentially glycosylated by GtfA/B, Nss, and Gly and then transferred to the Asp1/2/3 complex in which Asp1 mediates the interaction of the Asp1/2/3 complex with the lipid bilayer for targeting of matured GspB to the export machinery.
Organizational Affiliation:
From the Department of Cell Biology, Harvard Medical School, Boston, Massachusetts 02115.