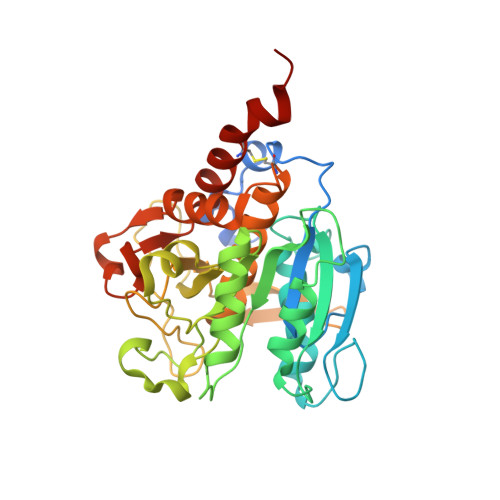
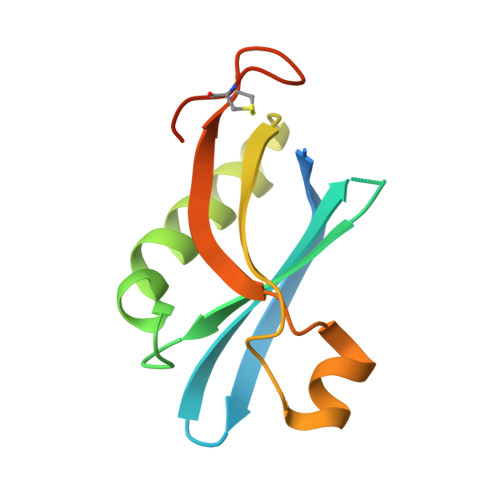
Structural Basis of Subtilase Cytotoxin Subab Assembly.
Le Nours, J., Paton, A.W., Byres, E., Troy, S., Herdman, B.P., Johnson, M.D., Paton, J.C., Rossjohn, J., Beddoe, T.(2013) J Biol Chem 288: 27505
- PubMed: 23921389
- DOI: https://doi.org/10.1074/jbc.M113.462622
- Primary Citation of Related Structures:
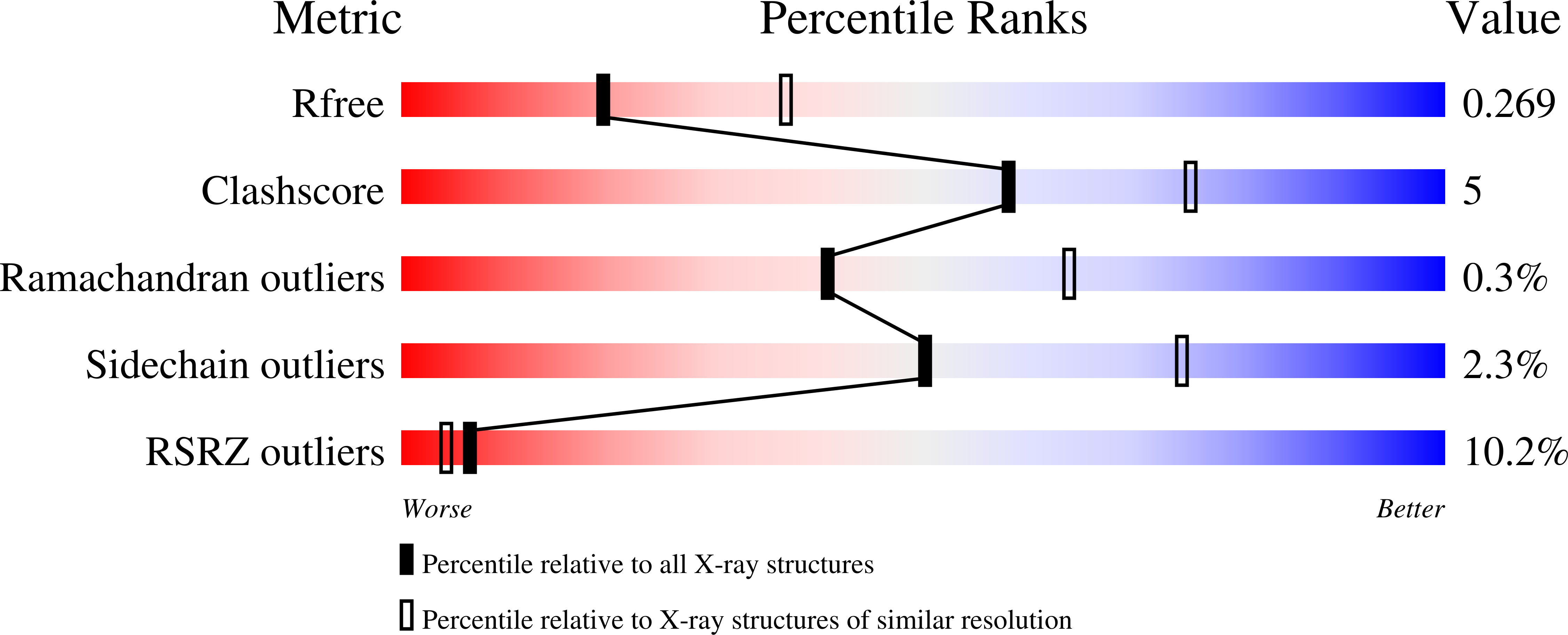
4BWG - PubMed Abstract:
Pathogenic strains of Escherichia coli produce a number of toxins that belong to the AB5 toxin family, which comprise a catalytic A-subunit that induces cellular dysfunction and a B-pentamer that recognizes host glycans. Although the molecular actions of many of the individual subunits of AB5 toxins are well understood, how they self-associate and the effect of this association on cytotoxicity are poorly understood. Here we have solved the structure of the holo-SubAB toxin that, in contrast to other AB5 toxins whose molecular targets are located in the cytosol, cleaves the endoplasmic reticulum chaperone BiP. SubA interacts with SubB in a similar manner to other AB5 toxins via the A2 helix and a conserved disulfide bond that joins the A1 domain with the A2 helix. The structure revealed that the active site of SubA is not occluded by the B-pentamer, and the B-pentamer does not enhance or inhibit the activity of SubA. Structure-based sequence comparisons with other AB5 toxin family members, combined with extensive mutagenesis studies on SubB, show how the hydrophobic patch on top of the B-pentamer plays a dominant role in binding the A-subunit. The structure of SubAB and the accompanying functional characterization of various mutants of SubAB provide a framework for understanding the important role of the B-pentamer in the assembly and the intracellular trafficking of this AB5 toxin.
Organizational Affiliation:
Australian Research Council (ARC) Centre of Excellence in Structural and Functional Microbial Genomics, Monash University, Clayton, Victoria 3800, Australia; Department of Biochemistry and Molecular Biology, School of Biomedical Sciences, Monash University, Clayton, Victoria 3800, Australia.