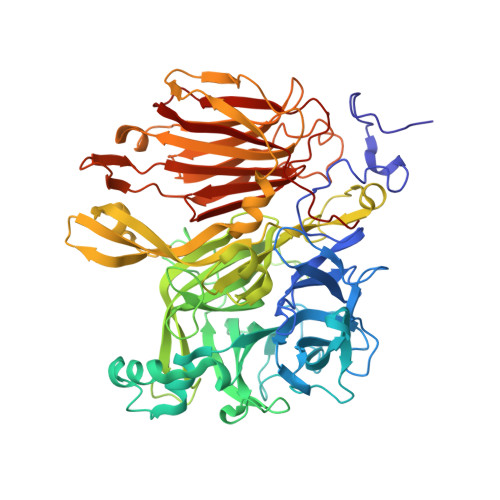
Crystal structure of Aspergillus japonicus fructosyltransferase complex with donor/acceptor substrates reveal complete sbusites in the active site for catalysis
Chuankhayan, P., Hsieh, C.Y., Huang, Y.C., Hsieh, Y.Y., Guan, H.H., Hsieh, Y.C., Tien, Y.C., Chen, C.D., Chaing, C.M., Chen, C.J.To be published.