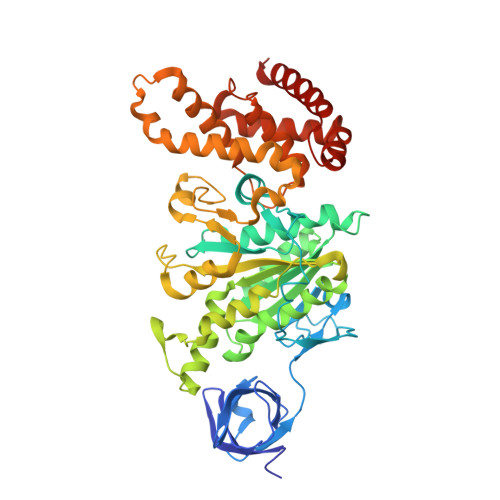
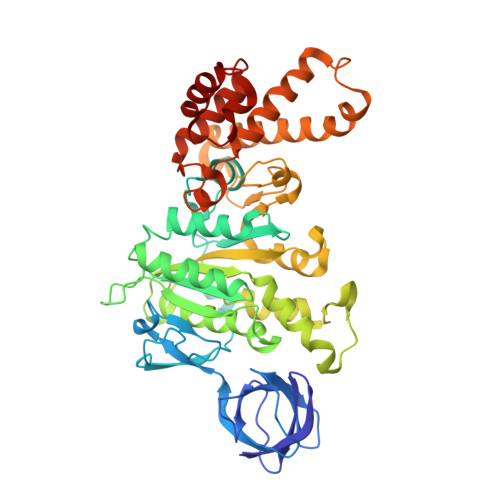
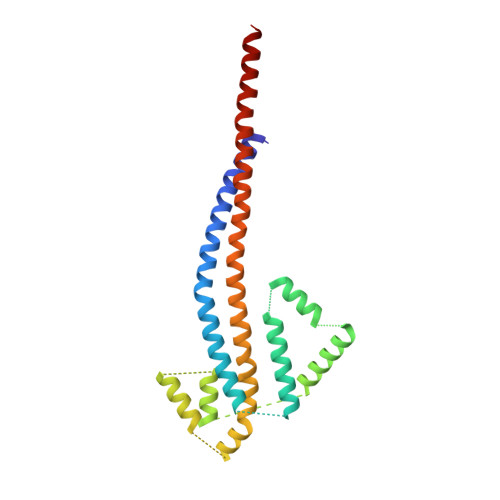
Structural features of the gamma subunit of the Escherichia coli F(1) ATPase revealed by a 4.4-A resolution map obtained by x-ray crystallography.
Hausrath, A.C., Gruber, G., Matthews, B.W., Capaldi, R.A.(1999) Proc Natl Acad Sci U S A 96: 13697-13702
- PubMed: 10570135
- DOI: https://doi.org/10.1073/pnas.96.24.13697
- Primary Citation of Related Structures:
1D8S - PubMed Abstract:
The F(1) part of the F(1)F(O) ATP synthase from Escherichia coli has been crystallized and its structure determined to 4.4-A resolution by using molecular replacement based on the structure of the beef-heart mitochondrial enzyme. The bacterial F(1) consists of five subunits with stoichiometry alpha(3), beta(3), gamma, delta, and epsilon. delta was removed before crystallization. In agreement with the structure of the beef-heart mitochondrial enzyme, although not that from rat liver, the present study suggests that the alpha and beta subunits are arranged in a hexagonal barrel but depart from exact 3-fold symmetry. In the structures of both beef heart and rat-liver mitochondrial F(1), less than half of the structure of the gamma subunit was seen because of presumed disorder in the crystals. The present electron-density map includes a number of rod-shaped features which appear to correspond to additional alpha-helical regions within the gamma subunit. These suggest that the gamma subunit traverses the full length of the stalk that links the F(1) and F(O) parts and makes significant contacts with the c subunit ring of F(O).
Organizational Affiliation:
Institute of Molecular Biology, Howard Hughes Medical Institute, Department of Physics, 1229 University of Oregon, Eugene, OR 97403-1229, USA.