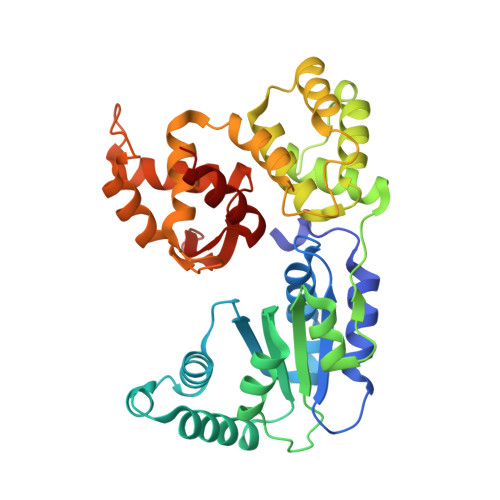
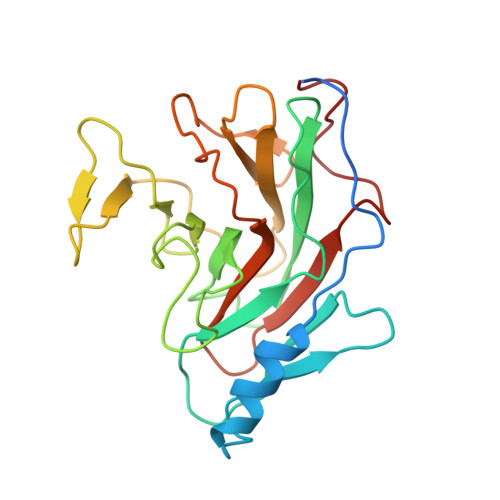
Resurrection of plant disease resistance proteins via helper NLR bioengineering.
Contreras, M.P., Pai, H., Selvaraj, M., Toghani, A., Lawson, D.M., Tumtas, Y., Duggan, C., Yuen, E.L.H., Stevenson, C.E.M., Harant, A., Maqbool, A., Wu, C.H., Bozkurt, T.O., Kamoun, S., Derevnina, L.(2023) Sci Adv 9: eadg3861-eadg3861
- PubMed: 37134163
- DOI: https://doi.org/10.1126/sciadv.adg3861
- Primary Citation of Related Structures:
8BV0 - PubMed Abstract:
Parasites counteract host immunity by suppressing helper nucleotide binding and leucine-rich repeat (NLR) proteins that function as central nodes in immune receptor networks. Understanding the mechanisms of immunosuppression can lead to strategies for bioengineering disease resistance. Here, we show that a cyst nematode virulence effector binds and inhibits oligomerization of the helper NLR protein NRC2 by physically preventing intramolecular rearrangements required for activation. An amino acid polymorphism at the binding interface between NRC2 and the inhibitor is sufficient for this helper NLR to evade immune suppression, thereby restoring the activity of multiple disease resistance genes. This points to a potential strategy for resurrecting disease resistance in crop genomes.
Organizational Affiliation:
The Sainsbury Laboratory, University of East Anglia, Norwich, UK.