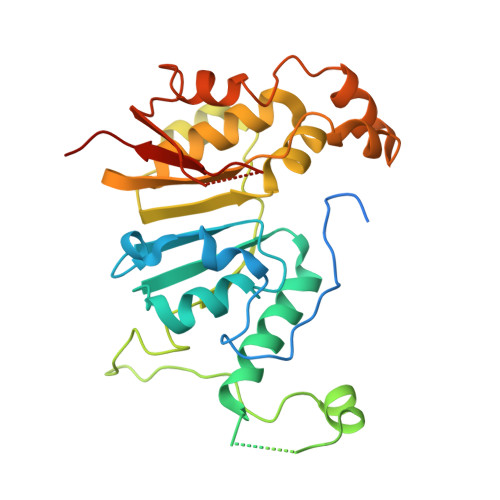
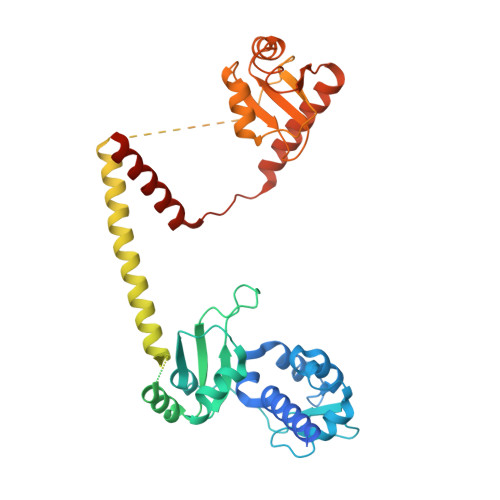
Structural basis of RNA conformational switching in the transcriptional regulator 7SK RNP.
Yang, Y., Liu, S., Egloff, S., Eichhorn, C.D., Hadjian, T., Zhen, J., Kiss, T., Zhou, Z.H., Feigon, J.(2022) Mol Cell 82: 1724
- PubMed: 35320752
- DOI: https://doi.org/10.1016/j.molcel.2022.03.001
- Primary Citation of Related Structures:
7SLP, 7SLQ - PubMed Abstract:
7SK non-coding RNA (7SK) negatively regulates RNA polymerase II (RNA Pol II) elongation by inhibiting positive transcription elongation factor b (P-TEFb), and its ribonucleoprotein complex (RNP) is hijacked by HIV-1 for viral transcription and replication. Methylphosphate capping enzyme (MePCE) and La-related protein 7 (Larp7) constitutively associate with 7SK to form a core RNP, while P-TEFb and other proteins dynamically assemble to form different complexes. Here, we present the cryo-EM structures of 7SK core RNP formed with two 7SK conformations, circular and linear, and uncover a common RNA-dependent MePCE-Larp7 complex. Together with NMR, biochemical, and cellular data, these structures reveal the mechanism of MePCE catalytic inactivation in the core RNP, unexpected interactions between Larp7 and RNA that facilitate a role as an RNP chaperone, and that MePCE-7SK-Larp7 core RNP serves as a scaffold for switching between different 7SK conformations essential for RNP assembly and regulation of P-TEFb sequestration and release.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of California, Los Angeles, Los Angeles, CA 90095, USA.