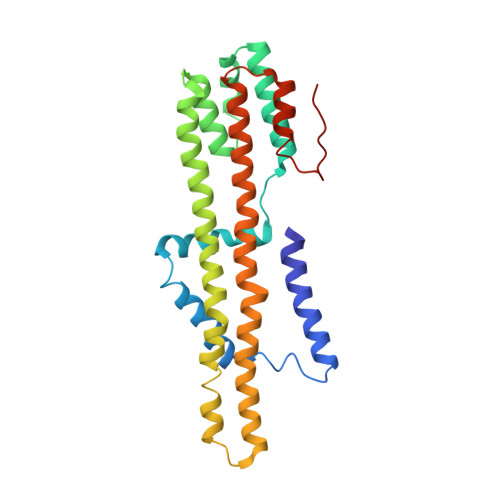
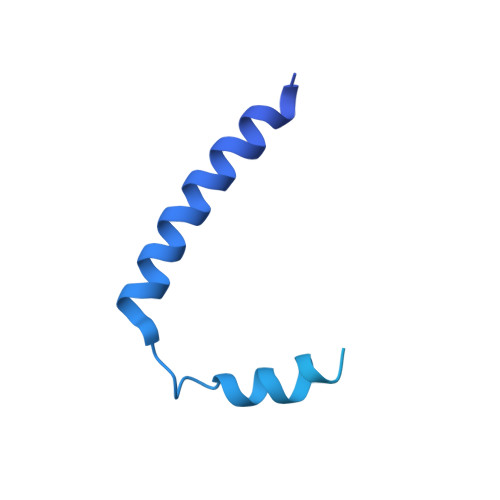
Structure and Function of Stator Units of the Bacterial Flagellar Motor.
Santiveri, M., Roa-Eguiara, A., Kuhne, C., Wadhwa, N., Hu, H., Berg, H.C., Erhardt, M., Taylor, N.M.I.(2020) Cell 183: 244-257.e16
- PubMed: 32931735
- DOI: https://doi.org/10.1016/j.cell.2020.08.016
- Primary Citation of Related Structures:
6YKM, 6YKP, 6YKR - PubMed Abstract:
Many bacteria use the flagellum for locomotion and chemotaxis. Its bidirectional rotation is driven by a membrane-embedded motor, which uses energy from the transmembrane ion gradient to generate torque at the interface between stator units and rotor. The structural organization of the stator unit (MotAB), its conformational changes upon ion transport, and how these changes power rotation of the flagellum remain unknown. Here, we present ~3 Å-resolution cryoelectron microscopy reconstructions of the stator unit in different functional states. We show that the stator unit consists of a dimer of MotB surrounded by a pentamer of MotA. Combining structural data with mutagenesis and functional studies, we identify key residues involved in torque generation and present a detailed mechanistic model for motor function and switching of rotational direction.
Organizational Affiliation:
Structural Biology of Molecular Machines Group, Protein Structure & Function Program, Novo Nordisk Foundation Center for Protein Research, Faculty of Health and Medical Sciences, University of Copenhagen, Blegdamsvej 3B, 2200 Copenhagen, Denmark.