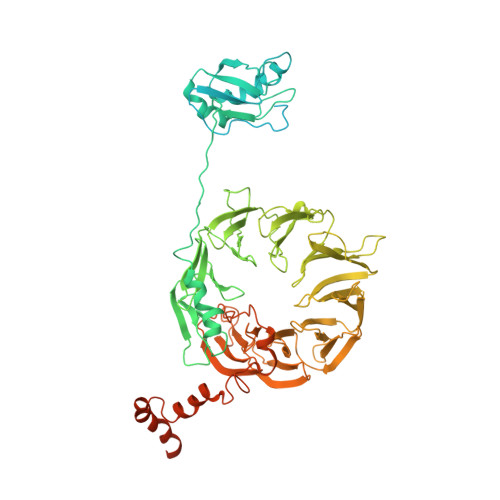
Structure of a human 48Stranslational initiation complex.
Brito Querido, J., Sokabe, M., Kraatz, S., Gordiyenko, Y., Skehel, J.M., Fraser, C.S., Ramakrishnan, V.(2020) Science 369: 1220-1227
- PubMed: 32883864
- DOI: https://doi.org/10.1126/science.aba4904
- Primary Citation of Related Structures:
6YBD, 6YBS, 6YBT, 6YBV, 6YBW, 6ZMW - PubMed Abstract:
A key step in translational initiation is the recruitment of the 43 S preinitiation complex by the cap-binding complex [eukaryotic initiation factor 4F (eIF4F)] at the 5' end of messenger RNA (mRNA) to form the 48 S initiation complex (i.e., the 48 S ). The 48 S then scans along the mRNA to locate a start codon. To understand the mechanisms involved, we used cryo-electron microscopy to determine the structure of a reconstituted human 48 S The structure reveals insights into early events of translation initiation complex assembly, as well as how eIF4F interacts with subunits of eIF3 near the mRNA exit channel in the 43 S The location of eIF4F is consistent with a slotting model of mRNA recruitment and suggests that downstream mRNA is unwound at least in part by being "pulled" through the 40 S subunit during scanning.
Organizational Affiliation:
MRC Laboratory of Molecular Biology, Cambridge, UK.