Increased versatility despite reduced molecular complexity: evolution, structure and function of metazoan splicing factor PRPF39.
De Bortoli, F., Neumann, A., Kotte, A., Timmermann, B., Schuler, T., Wahl, M.C., Loll, B., Heyd, F.(2019) Nucleic Acids Res 47: 5867-5879
- PubMed: 30949712
- DOI: https://doi.org/10.1093/nar/gkz243
- Primary Citation of Related Structures:
6G70 - PubMed Abstract:
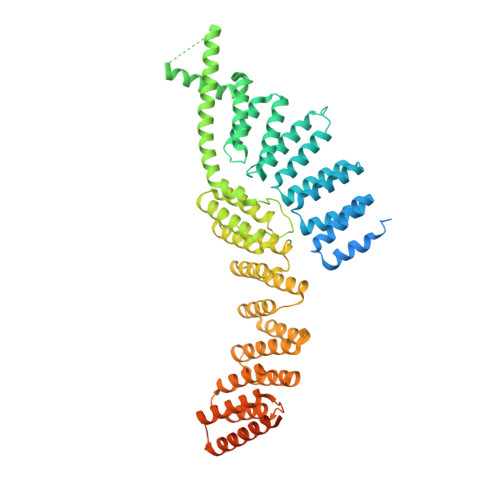
In the yeast U1 snRNP the Prp39/Prp42 heterodimer is essential for early steps of spliceosome assembly. In metazoans no Prp42 ortholog exists, raising the question how the heterodimer is functionally substituted. Here we present the crystal structure of murine PRPF39, which forms a homodimer. Structure-guided point mutations disrupt dimer formation and inhibit splicing, manifesting the homodimer as functional unit. PRPF39 expression is controlled by NMD-inducing alternative splicing in mice and human, suggesting a role in adapting splicing efficiency to cell type specific requirements. A phylogenetic analysis reveals coevolution of shortened U1 snRNA and the absence of Prp42, which correlates with overall splicing complexity in different fungi. While current models correlate the diversity of spliceosomal proteins with splicing complexity, our study highlights a contrary case. We find that organisms with higher splicing complexity have substituted the Prp39/Prp42 heterodimer with a PRPF39 homodimer.
Organizational Affiliation:
Institut für Chemie und Biochemie, RNA Biochemie, Freie Universität Berlin, Takustr. 6, 14195 Berlin, Germany.