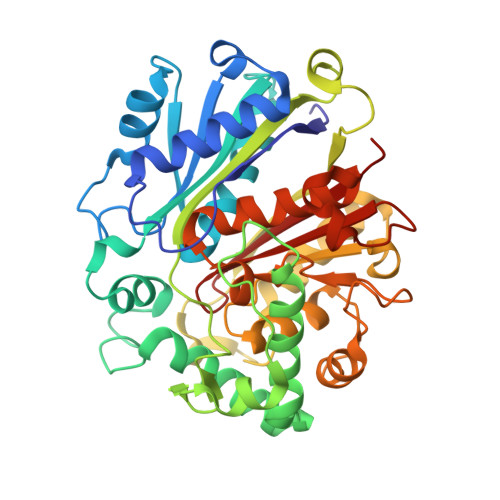
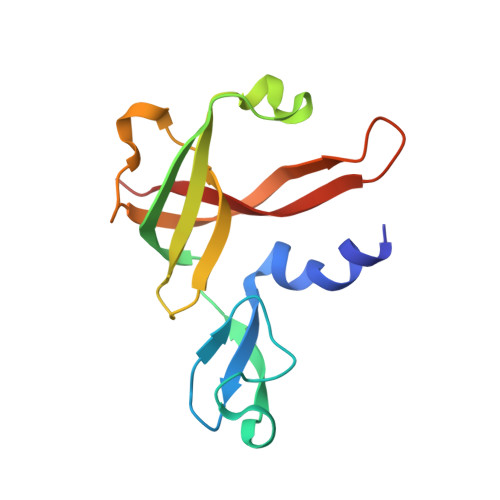
Archaeal acetoacetyl-CoA thiolase/HMG-CoA synthase complex channels the intermediate via a fused CoA-binding site.
Vogeli, B., Engilberge, S., Girard, E., Riobe, F., Maury, O., Erb, T.J., Shima, S., Wagner, T.(2018) Proc Natl Acad Sci U S A 115: 3380-3385
- PubMed: 29531083
- DOI: https://doi.org/10.1073/pnas.1718649115
- Primary Citation of Related Structures:
6ESQ, 6ET9 - PubMed Abstract:
Many reactions within a cell are thermodynamically unfavorable. To efficiently run some of those endergonic reactions, nature evolved intermediate-channeling enzyme complexes, in which the products of the first endergonic reactions are immediately consumed by the second exergonic reactions. Based on this concept, we studied how archaea overcome the unfavorable first reaction of isoprenoid biosynthesis-the condensation of two molecules of acetyl-CoA to acetoacetyl-CoA catalyzed by acetoacetyl-CoA thiolases (thiolases). We natively isolated an enzyme complex comprising the thiolase and 3-hydroxy-3-methylglutaryl (HMG)-CoA synthase (HMGCS) from a fast-growing methanogenic archaeon, Methanothermococcus thermolithotrophicus HMGCS catalyzes the second reaction in the mevalonate pathway-the exergonic condensation of acetoacetyl-CoA and acetyl-CoA to HMG-CoA. The 380-kDa crystal structure revealed that both enzymes are held together by a third protein (DUF35) with so-far-unknown function. The active-site clefts of thiolase and HMGCS form a fused CoA-binding site, which allows for efficient coupling of the endergonic thiolase reaction with the exergonic HMGCS reaction. The tripartite complex is found in almost all archaeal genomes and in some bacterial ones. In addition, the DUF35 proteins are also important for polyhydroxyalkanoate (PHA) biosynthesis, most probably by functioning as a scaffold protein that connects thiolase with 3-ketoacyl-CoA reductase. This natural and highly conserved enzyme complex offers great potential to improve isoprenoid and PHA biosynthesis in biotechnologically relevant organisms.
Organizational Affiliation:
Biochemistry and Synthetic Metabolism, Max Planck Institute for Terrestrial Microbiology, 35043 Marburg, Germany.