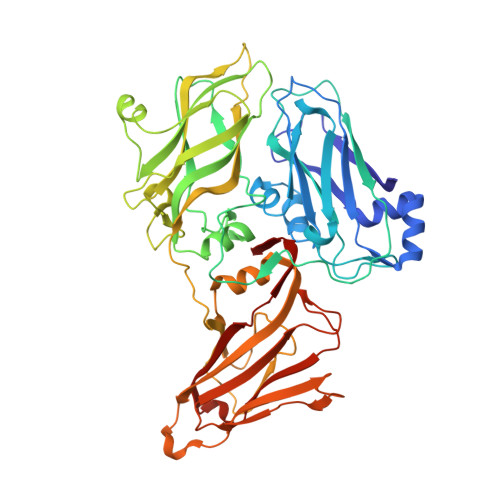
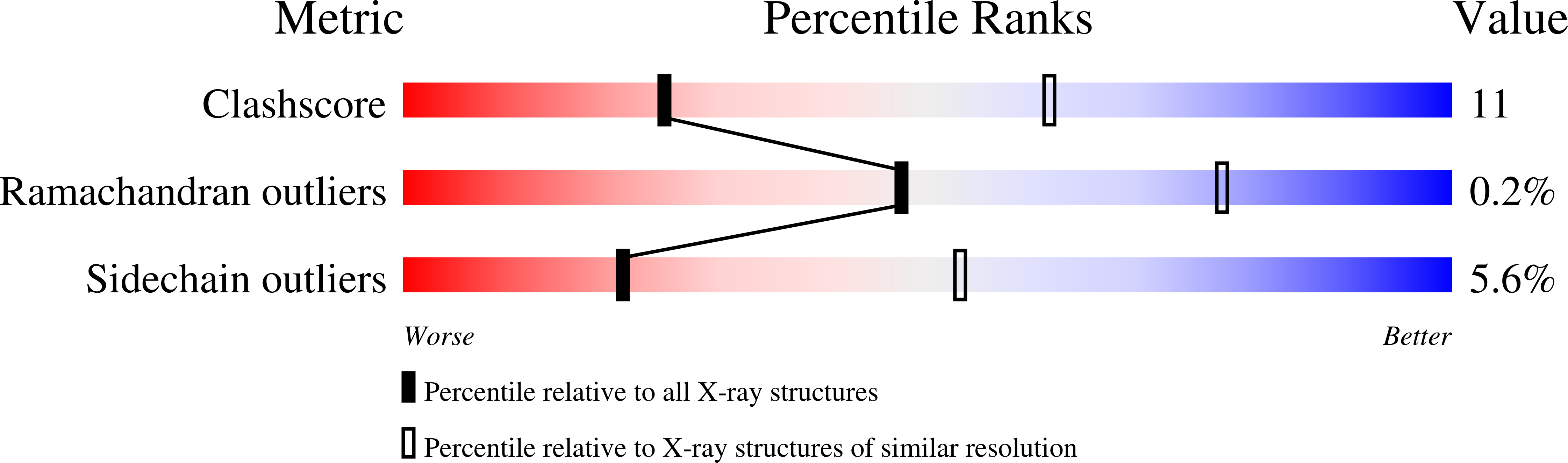Exploiting Protein Engineering and Crystal Polymorphism for Successful X-Ray Structure Determination
Bonnefond, L., Schellenberger, P., Basquin, J., Demangeat, G., Ritzenthaler, C., Chenevert, R., Balg, C., Frugier, M., Rudinger-Thirion, J., Giege, R., Lorber, B., Sauter, C.(2011) Cryst Growth Des 11: 4334