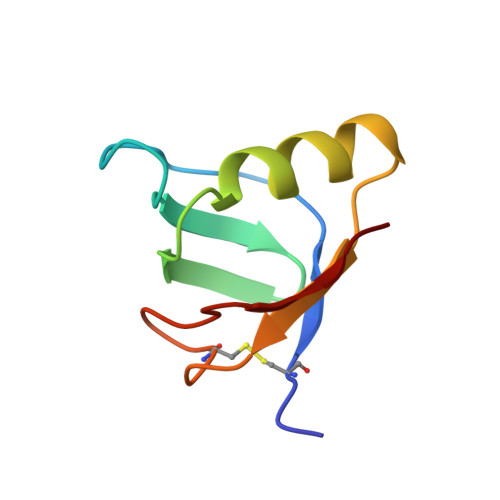
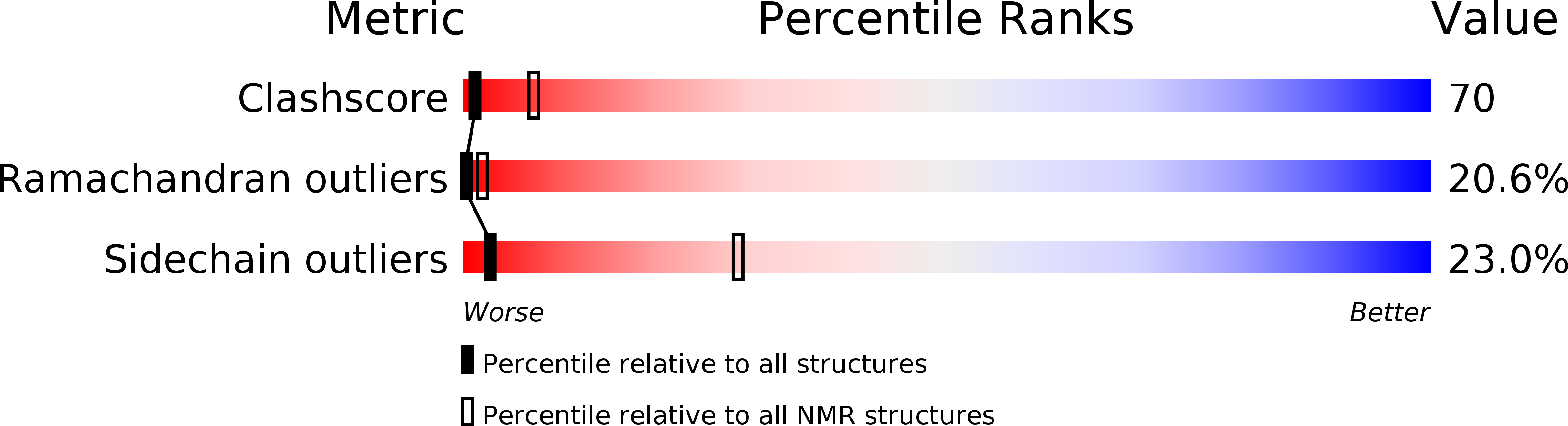Solution structure of the carbohydrate-binding B-subunit homopentamer of verotoxin VT-1 from E. coli.
Richardson, J.M., Evans, P.D., Homans, S.W., Donohue-Rolfe, A.(1997) Nat Struct Biol 4: 190-193
- PubMed: 9164458
- DOI: https://doi.org/10.1038/nsb0397-190
- Primary Citation of Related Structures:
4ULL