Molecular and Structural Basis for the Roles of Hepatitis C Virus Polymerase NS5B Amino Acids 15, 223, and 321 in Viral Replication and Drug Resistance.
Lam, A.M., Edwards, T.E., Mosley, R.T., Murakami, E., Bansal, S., Lugo, C., Bao, H., Otto, M.J., Sofia, M.J., Furman, P.A.(2014) Antimicrob Agents Chemother 58: 6861-6869
- PubMed: 25182647
- DOI: https://doi.org/10.1128/AAC.03847-14
- Primary Citation of Related Structures:
4OBC - PubMed Abstract:
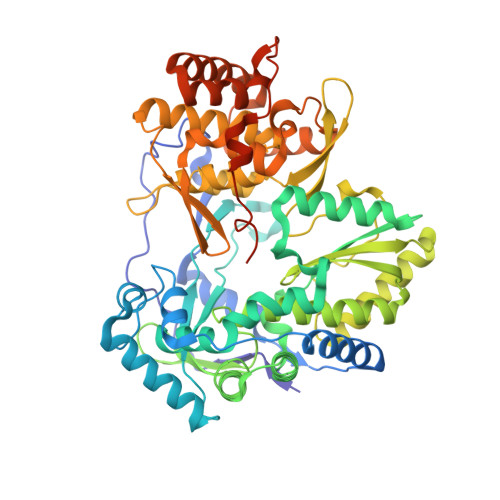
Resistance to the 2'-F-2'-C-methylguanosine monophosphate nucleotide hepatitis C virus (HCV) inhibitors PSI-352938 and PSI-353661 was associated with a combination of amino acid changes (changes of S to G at position 15 [S15G], C223H, and V321I) within the genotype 2a nonstructural protein 5B (NS5B), an RNA-dependent RNA polymerase. To understand the role of these residues in viral replication, we examined the effects of single and multiple point mutations on replication fitness and inhibition by a series of nucleotide analog inhibitors. An acidic residue at position 15 reduced replicon fitness, consistent with its proximity to the RNA template. A change of the residue at position 223 to an acidic or large residue reduced replicon fitness, consistent with its proposed proximity to the incoming nucleoside triphosphate (NTP). A change of the residue at position 321 to a charged residue was not tolerated, consistent with its position within a hydrophobic cavity. This triple resistance mutation was specific to both genotype 2a virus and 2'-F-2'-C-methylguanosine inhibitors. A crystal structure of the NS5B S15G/C223H/V321I mutant of the JFH-1 isolate exhibited rearrangement to a conformation potentially consistent with short primer-template RNA binding, which could suggest a mechanism of resistance accomplished through a change in the NS5B conformation, which was better tolerated by genotype 2a virus than by viruses of other genotypes.
Organizational Affiliation:
Pharmasset, Inc., Princeton, New Jersey, USA alam@noviratherapeutics.com tedwards@be4.com.