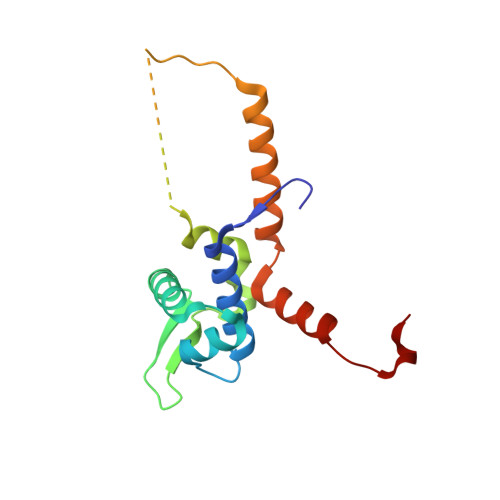
The Unique Regulation of Iron-Sulfur Cluster Biogenesis in a Gram-Positive Bacterium.
Santos, J.A., Alonso-Garcia, N., Macedo-Ribeiro, S., Pereira, P.J.B.(2014) Proc Natl Acad Sci U S A 111: E2251
- PubMed: 24847070
- DOI: https://doi.org/10.1073/pnas.1322728111
- Primary Citation of Related Structures:
4CHU, 4CIC - PubMed Abstract:
Iron-sulfur clusters function as cofactors of a wide range of proteins, with diverse molecular roles in both prokaryotic and eukaryotic cells. Dedicated machineries assemble the clusters and deliver them to the final acceptor molecules in a tightly regulated process. In the prototypical Gram-negative bacterium Escherichia coli, the two existing iron-sulfur cluster assembly systems, iron-sulfur cluster (ISC) and sulfur assimilation (SUF) pathways, are closely interconnected. The ISC pathway regulator, IscR, is a transcription factor of the helix-turn-helix type that can coordinate a [2Fe-2S] cluster. Redox conditions and iron or sulfur availability modulate the ligation status of the labile IscR cluster, which in turn determines a switch in DNA sequence specificity of the regulator: cluster-containing IscR can bind to a family of gene promoters (type-1) whereas the clusterless form recognizes only a second group of sequences (type-2). However, iron-sulfur cluster biogenesis in Gram-positive bacteria is not so well characterized, and most organisms of this group display only one of the iron-sulfur cluster assembly systems. A notable exception is the unique Gram-positive dissimilatory metal reducing bacterium Thermincola potens, where genes from both systems could be identified, albeit with a diverging organization from that of Gram-negative bacteria. We demonstrated that one of these genes encodes a functional IscR homolog and is likely involved in the regulation of iron-sulfur cluster biogenesis in T. potens. Structural and biochemical characterization of T. potens and E. coli IscR revealed a strikingly similar architecture and unveiled an unforeseen conservation of the unique mechanism of sequence discrimination characteristic of this distinctive group of transcription regulators.
Organizational Affiliation:
Instituto de Biologia Molecular e Celular (IBMC), Universidade do Porto, 4150-180 Porto, Portugal; andInstituto de Ciências Biomédicas de Abel Salazar (ICBAS), Universidade do Porto, 4050-313 Porto, Portugal.