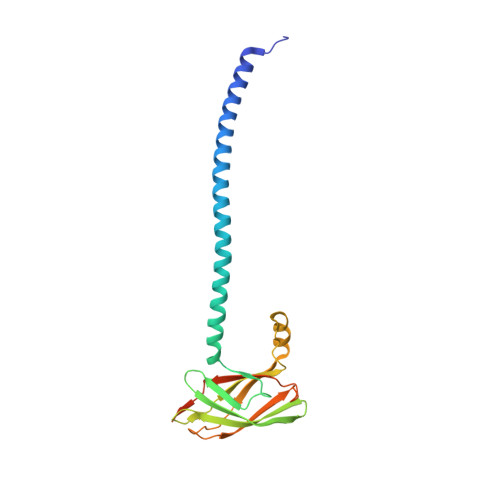
A Trimeric Lipoprotein Assists in Trimeric Autotransporter Biogenesis in Enterobacteria.
Grin, I., Hartmann, M.D., Sauer, G., Hernandez Alvarez, B., Schutz, M., Madlung, J., Macek, B., Felipe-Lopez, A., Hensel, M., Lupas, A., Linke, D.(2014) J Biol Chem 289: 7388
- PubMed: 24369174
- DOI: https://doi.org/10.1074/jbc.M113.513275
- Primary Citation of Related Structures:
4C47 - PubMed Abstract:
Trimeric autotransporter adhesins (TAAs) are important virulence factors of many Gram-negative bacterial pathogens. TAAs form fibrous, adhesive structures on the bacterial cell surface. Their N-terminal extracellular domains are exported through a C-terminal membrane pore; the insertion of the pore domain into the bacterial outer membrane follows the rules of β-barrel transmembrane protein biogenesis and is dependent on the essential Bam complex. We have recently described the full fiber structure of SadA, a TAA of unknown function in Salmonella and other enterobacteria. In this work, we describe the structure and function of SadB, a small inner membrane lipoprotein. The sadB gene is located in an operon with sadA; orthologous operons are only found in enterobacteria, whereas other TAAs are not typically associated with lipoproteins. Strikingly, SadB is also a trimer, and its co-expression with SadA has a direct influence on SadA structural integrity. This is the first report of a specific export factor of a TAA, suggesting that at least in some cases TAA autotransport is assisted by additional periplasmic proteins.
Organizational Affiliation:
From the Max Planck Institut für Entwicklungsbiologie, 72076 Tübingen.