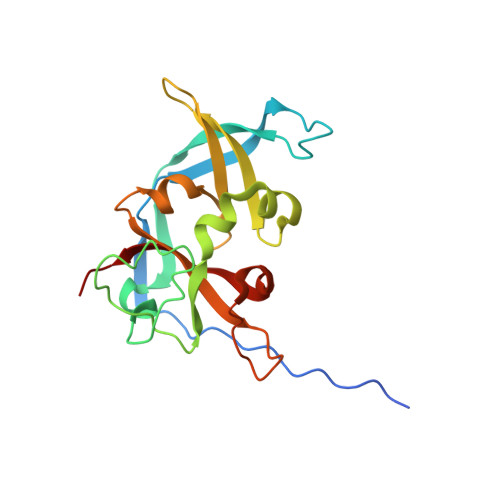
Crystal structure of the read-through domain from bacteriophage Qbeta A1 protein
Rumnieks, J., Tars, K.(2011) Protein Sci 20: 1707-1712
- PubMed: 21805520
- DOI: https://doi.org/10.1002/pro.704
- Primary Citation of Related Structures:
3RLC, 3RLK - PubMed Abstract:
Bacteriophage Qβ is a small RNA virus that infects Escherichia coli. The virus particle contains a few copies of the minor coat protein A1, a C-terminally prolonged version of the coat protein, which is formed when ribosomes occasionally read-through the leaky stop codon of the coat protein. The crystal structure of the read-through domain from bacteriophage Qβ A1 protein was determined at a resolution of 1.8 Å. The domain consists of a heavily deformed five-stranded β-barrel on one side of the protein and a β-hairpin and a three-stranded β-sheet on the other. Several short helices and well-ordered loops are also present throughout the protein. The N-terminal part of the read-through domain contains a prominent polyproline type II helix. The overall fold of the domain is not similar to any published structure in the Protein Data Bank.
Organizational Affiliation:
Latvian Biomedical Research and Study Centre, Ratsupites 1, Riga LV-1067, Latvia. j.rumnieks@biomed.lu.lv