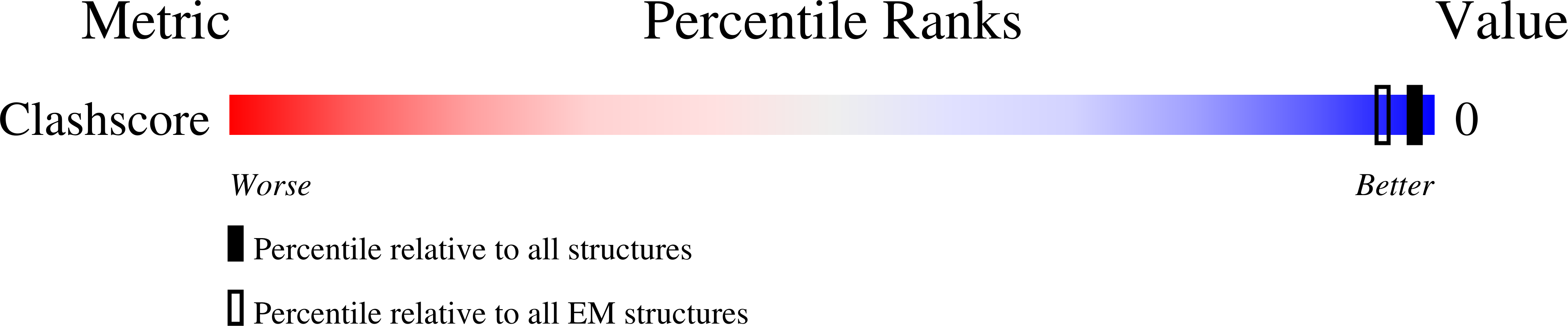Structural studies of the sputnik virophage.
Sun, S., La Scola, B., Bowman, V.D., Ryan, C.M., Whitelegge, J.P., Raoult, D., Rossmann, M.G.(2010) J Virol 84: 894-897
- PubMed: 19889775
- DOI: https://doi.org/10.1128/JVI.01957-09
- Primary Citation of Related Structures:
3KK5 - PubMed Abstract:
The virophage Sputnik is a satellite virus of the giant mimivirus and is the only satellite virus reported to date whose propagation adversely affects its host virus' production. Genome sequence analysis showed that Sputnik has genes related to viruses infecting all three domains of life. Here, we report structural studies of Sputnik, which show that it is about 740 A in diameter, has a T=27 icosahedral capsid, and has a lipid membrane inside the protein shell. Structural analyses suggest that the major capsid protein of Sputnik is likely to have a double jelly-roll fold, although sequence alignments do not show any detectable similarity with other viral double jelly-roll capsid proteins. Hence, the origin of Sputnik's capsid might have been derived from other viruses prior to its association with mimivirus.
Organizational Affiliation:
Department of Biological Sciences, Purdue University, 915 W. State Street, West Lafayette, IN 47907-2054, USA.