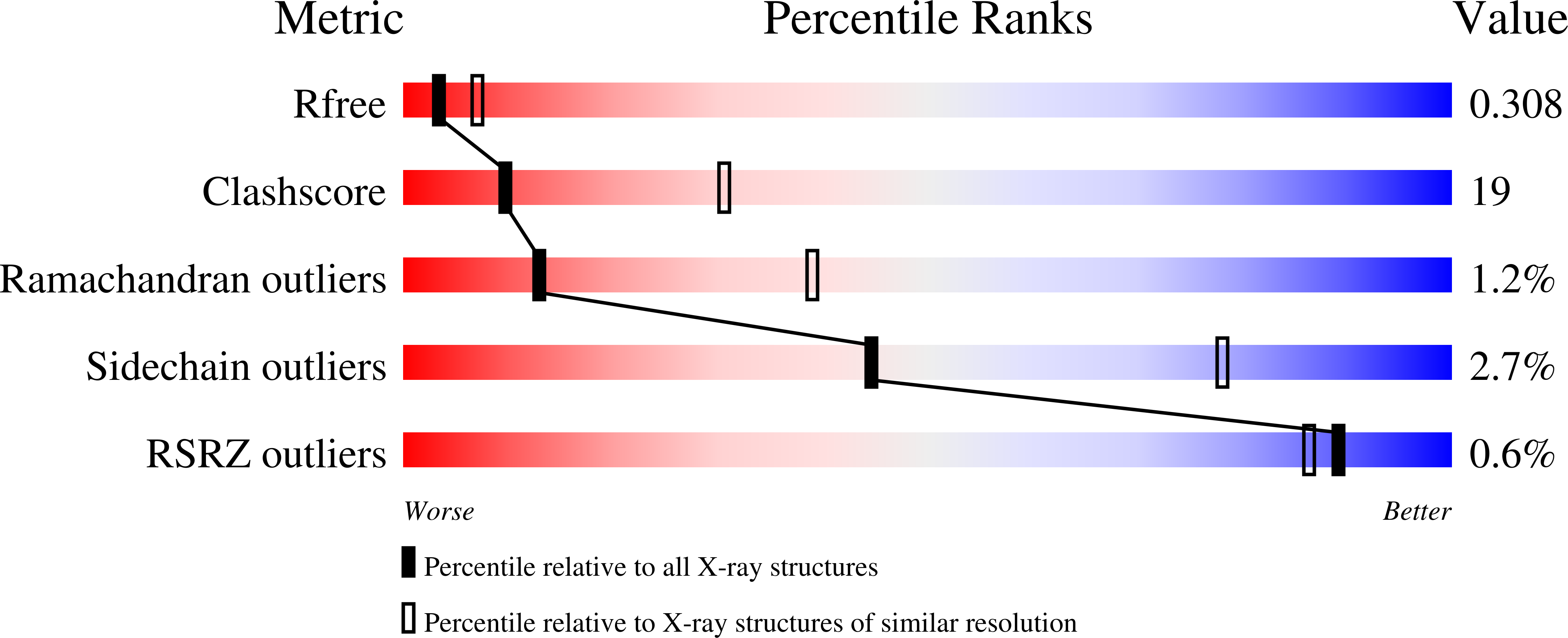
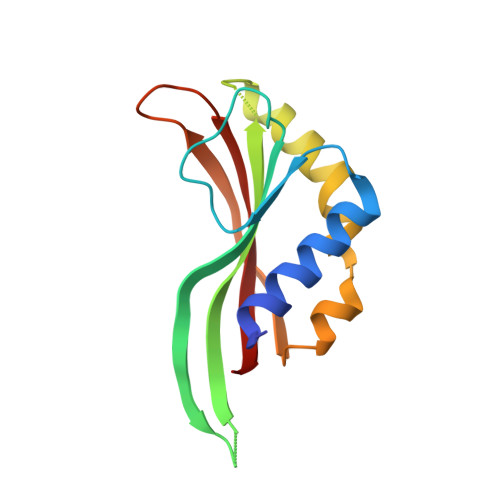
The X-ray crystal structure of the phage lambda tail terminator protein reveals the biologically relevant hexameric ring structure and demonstrates a conserved mechanism of tail termination among diverse long-tailed phages.
Pell, L.G., Liu, A., Edmonds, L., Donaldson, L.W., Howell, P.L., Davidson, A.R.(2009) J Mol Biol 389: 938-951
- PubMed: 19426744
- DOI: https://doi.org/10.1016/j.jmb.2009.04.072
- Primary Citation of Related Structures:
3FZ2, 3FZB - PubMed Abstract:
The tail terminator protein (TrP) plays an essential role in phage tail assembly by capping the rapidly polymerizing tail once it has reached its requisite length and serving as the interaction surface for phage heads. Here, we present the 2.7-A crystal structure of a hexameric ring of gpU, the TrP of phage lambda. Using sequence alignment analysis and site-directed mutagenesis, we have shown that this multimeric structure is biologically relevant and we have delineated its functional surfaces. Comparison of the hexameric crystal structure with the solution structure of gpU that we previously solved using NMR spectroscopy shows large structural changes occurring upon multimerization and suggests a mechanism that allows gpU to remain monomeric at high concentrations on its own, yet polymerize readily upon contact with an assembled tail tube. The gpU hexamer displays several flexible loops that play key roles in head and tail binding, implying a role for disorder-to-order transitions in controlling assembly as has been observed with other lambda morphogenetic proteins. Finally, we have found that the hexameric structure of gpU is very similar to the structure of a putative TrP from a contractile phage tail even though it displays no detectable sequence similarity. This finding coupled with further bioinformatic investigations has led us to conclude that the TrPs of non-contractile-tailed phages, such as lambda, are evolutionarily related to those of contractile-tailed phages, such as P2 and Mu, and that all long-tailed phages may utilize a conserved mechanism for tail termination.
Organizational Affiliation:
Department of Biochemistry, University of Toronto, ON, Canada.