Crystal structure of native RPE65, the retinoid isomerase of the visual cycle.
Kiser, P.D., Golczak, M., Lodowski, D.T., Chance, M.R., Palczewski, K.(2009) Proc Natl Acad Sci U S A 106: 17325-17330
- PubMed: 19805034
- DOI: https://doi.org/10.1073/pnas.0906600106
- Primary Citation of Related Structures:
3FSN - PubMed Abstract:
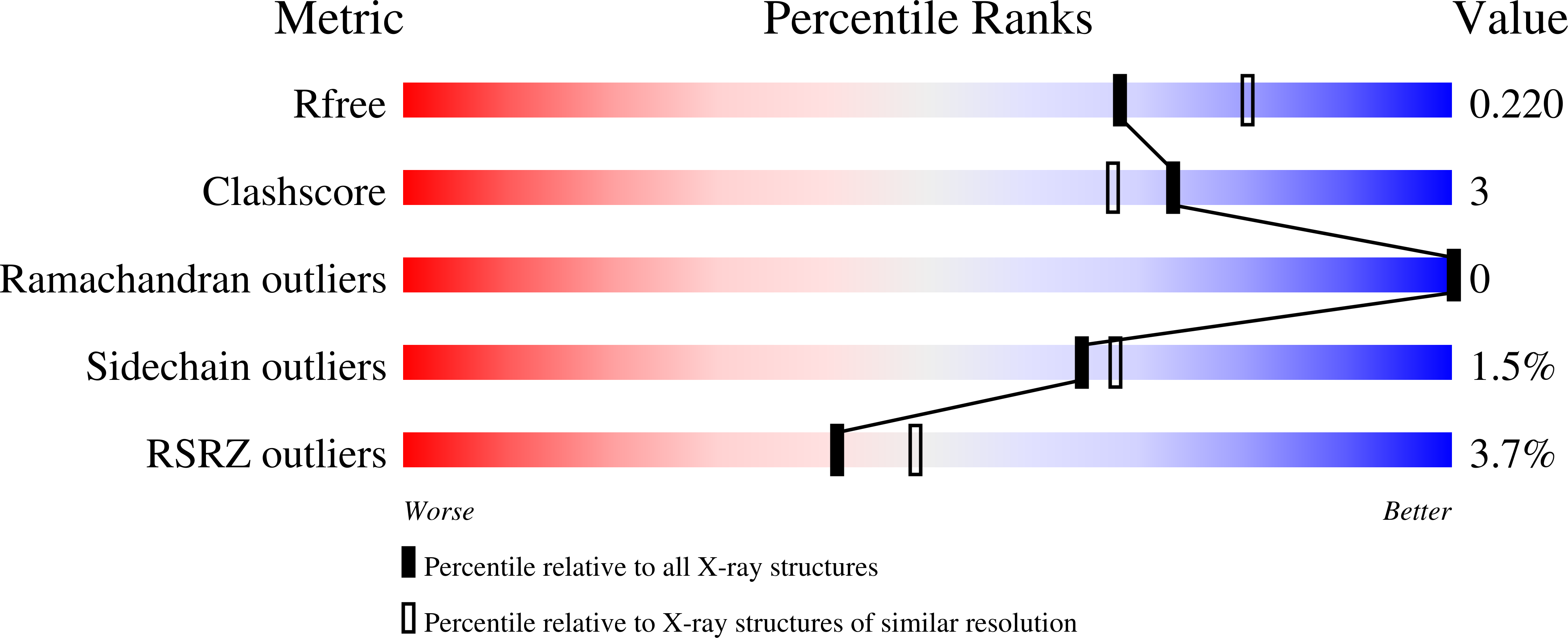
Vertebrate vision is maintained by the retinoid (visual) cycle, a complex enzymatic pathway that operates in the retina to regenerate the visual chromophore, 11-cis-retinal. A key enzyme in this pathway is the microsomal membrane protein RPE65. This enzyme catalyzes the conversion of all-trans-retinyl esters to 11-cis-retinol in the retinal pigment epithelium (RPE). Mutations in RPE65 are known to be responsible for a subset of cases of the most common form of childhood blindness, Leber congenital amaurosis (LCA). Although retinoid isomerase activity has been attributed to RPE65, its catalytic mechanism remains a matter of debate. Also, the manner in which RPE65 binds to membranes and extracts retinoid substrates is unclear. To gain insight into these questions, we determined the crystal structure of native bovine RPE65 at 2.14-A resolution. The structural, biophysical, and biochemical data presented here provide the framework needed for an in-depth understanding of the mechanism of catalytic isomerization and membrane association, in addition to the role mutations that cause LCA have in disrupting protein function.
Organizational Affiliation:
Department of Pharmacology, School of Medicine, Case Western Reserve University, Cleveland, OH 44106-4965, USA.