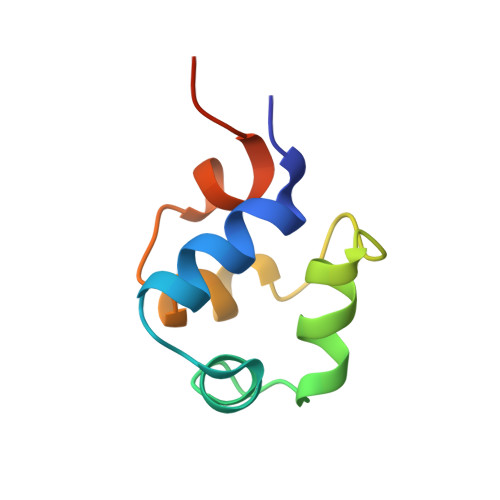
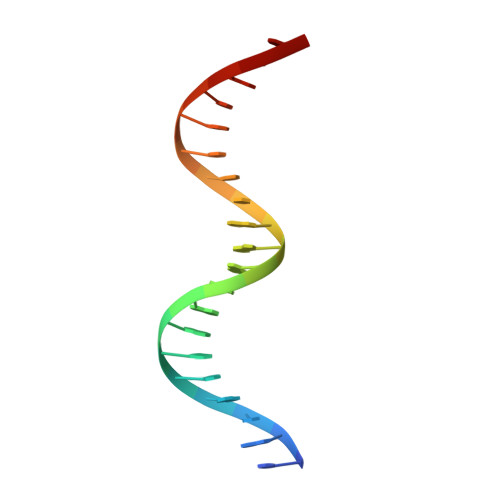
The phage 434 Cro/OR1 complex at 2.5 A resolution.
Mondragon, A., Harrison, S.C.(1991) J Mol Biol 219: 321-334
- PubMed: 2038059
- DOI: https://doi.org/10.1016/0022-2836(91)90568-q
- Primary Citation of Related Structures:
3CRO - PubMed Abstract:
The crystal structure of phage 434 Cro protein in complex with a 20 base-pair DNA fragment has been determined to 2.5 A resolution. The DNA fragment contains the sequence of the OR1 operator site. The structure shows a bent conformation for the DNA, straighter at the center and more bent at the ends. The central base-pairs adopt conformations with significant deviations from coplanarity. The two molecules interact extensively along their common interface, both through hydrogen bonds and van der Waals interactions. The significance of these interactions for operator binding and recognition is discussed.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Harvard University, Cambridge, MA 02138.