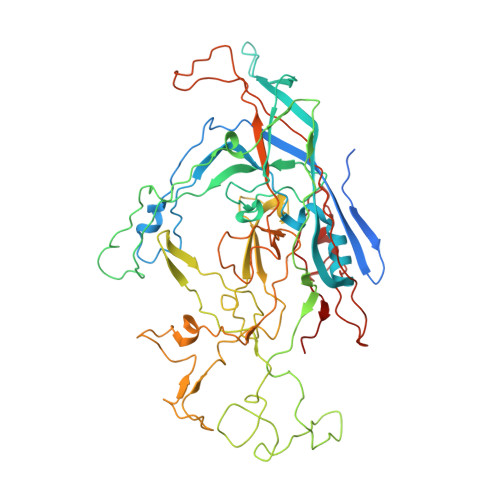
Structure of a Packaging-Defective Mutant of Minute Virus of Mice Indicates that the Genome is Packaged Via a Pore at a 5-Fold Axis.
Plevka, P., Hafenstein, S., Li, L., D'Abrgamo, A., Cotmore, S.F., Rossmann, M.G., Tattersall, P.(2011) J Virol 85: 4822
- PubMed: 21367911
- DOI: https://doi.org/10.1128/JVI.02598-10
- Primary Citation of Related Structures:
2XGK - PubMed Abstract:
The parvovirus minute virus of mice (MVM) packages a single copy of its linear single-stranded DNA genome into preformed capsids, in a process that is probably driven by a virus-encoded helicase. Parvoviruses have a roughly cylindrically shaped pore that surrounds each of the 12 5-fold vertices. The pore, which penetrates the virion shell, is created by the juxtaposition of 10 antiparallel β-strands, two from each of the 5-fold-related capsid proteins. There is a bottleneck in the channel formed by the symmetry-related side chains of the leucines at position 172. We report here the X-ray crystal structure of the particles produced by a leucine-to-tryptophan mutation at position 172 and the analysis of its biochemical properties. The mutant capsid had its 5-fold channel blocked, and the particles were unable to package DNA, strongly suggesting that the 5-fold pore is the packaging portal for genome entry.
Organizational Affiliation:
Department of Biological Sciences, Purdue University, West Lafayette, Indiana 47907, USA.