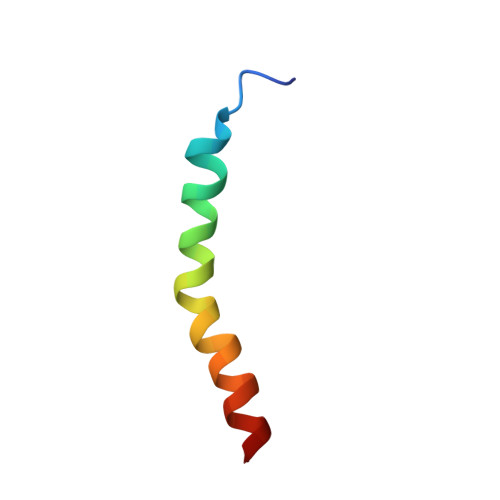
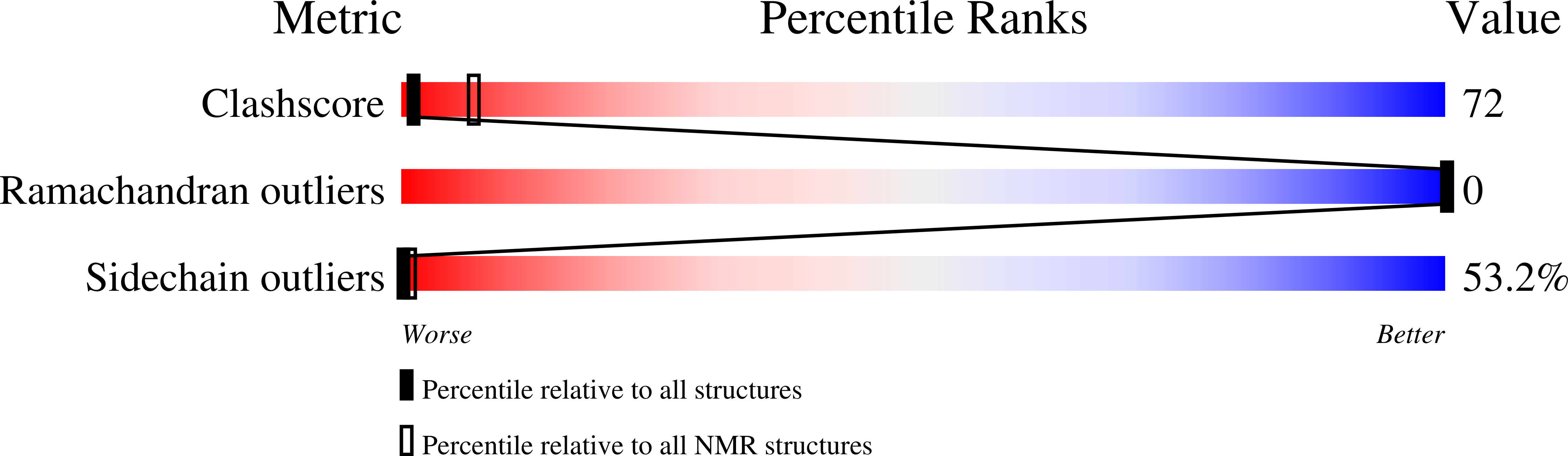Three-dimensional structure of the two peptides that constitute the two-peptide bacteriocin plantaricin EF
Fimland, N., Rogne, P., Fimland, G., Nissen-Meyer, J., Kristiansen, P.E.(2008) Biochim Biophys Acta 1784: 1711-1719
- PubMed: 18555030
- DOI: https://doi.org/10.1016/j.bbapap.2008.05.003
- Primary Citation of Related Structures:
2JUI, 2RLW - PubMed Abstract:
The three-dimensional structures of the two peptides plantaricin E (plnE; 33 residues) and plantaricin F (plnF; 34 residues) constituting the two-peptide bacteriocin plantaricin EF (plnEF) have been determined by nuclear magnetic resonance (NMR) spectroscopy in the presence of DPC micelles. PlnE has an N-terminal alpha-helix (residues 10-21), and a C-terminal alpha-helix-like structure (residues 25-31). PlnF has a long central alpha-helix (residues 7-32) with a kink of 38+/-7 degrees at Pro20. There is some flexibility in the helix in the kink region. Both helices in plnE are amphiphilic, while the helix in plnF is polar in its N-terminal half and amphiphilic in its C-terminal half. The alpha-helical content obtained by NMR spectroscopy is in agreement with CD studies. PlnE has two GxxxG motifs which are putative helix-helix interaction motifs, one at residues 5 to 9 and one at residues 20 to 24, while plnF has one such motif at residues 30 to 34. The peptides are flexible in these GxxxG regions. It is suggested that the two peptides lie parallel in a staggered fashion relative to each other and interact through helix-helix interactions involving the GxxxG motifs.
Organizational Affiliation:
Department of Molecular Biosciences, University of Oslo, Pb. 1041 Blindern, 0316 Oslo, Norway.