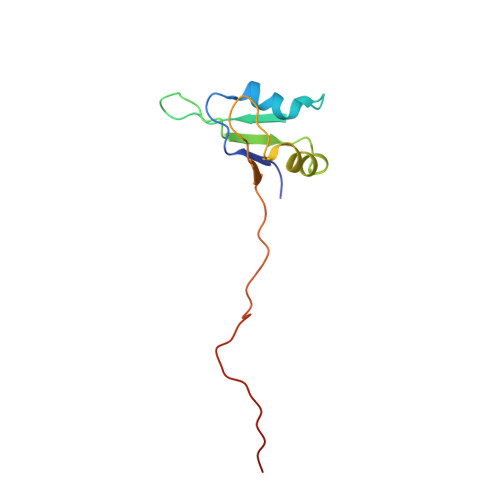
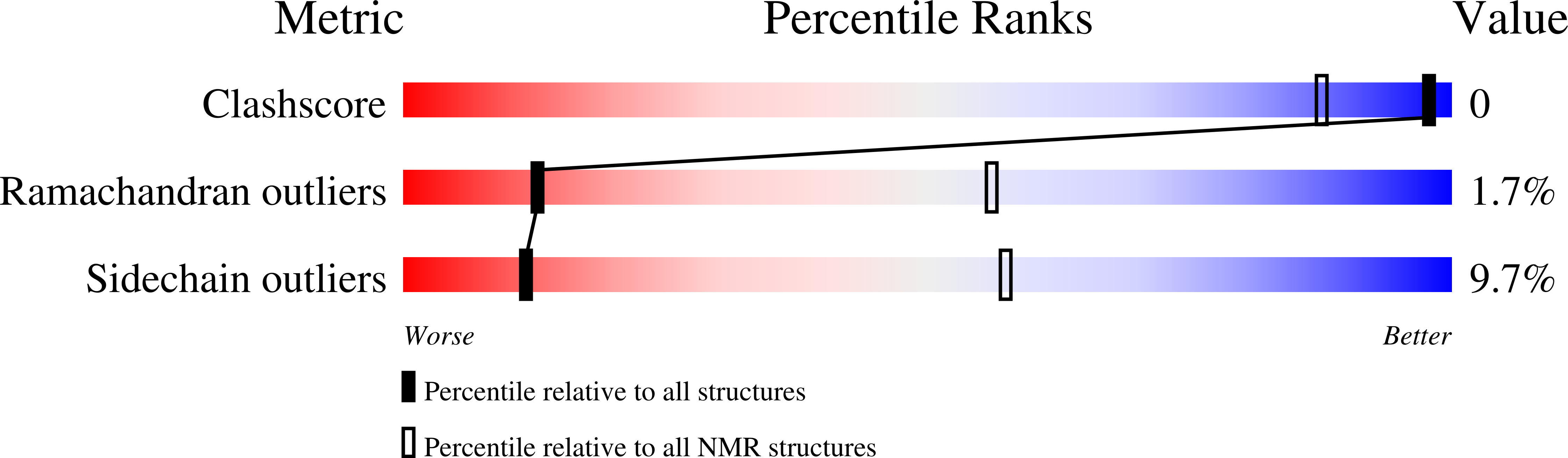Gbp2 interacts with THO/TREX through a novel type of RRM domain.
Martinez-Lumbreras, S., Taverniti, V., Zorrilla, S., Seraphin, B., Perez-Canadillas, J.M.(2016) Nucleic Acids Res 44: 437-448
- PubMed: 26602689
- DOI: https://doi.org/10.1093/nar/gkv1303
- Primary Citation of Related Structures:
2MZQ, 2MZR, 2MZS, 2MZT - PubMed Abstract:
Metazoan SR and SR-like proteins are important regulatory factors in RNA splicing, export, translation and RNA decay. We determined the NMR structures and nucleic acid interaction modes of Gbp2 and Hrb1, two paralogous budding yeast proteins with similarities to mammalian SR proteins. Gbp2 RRM1 and RRM2 recognise preferentially RNAs containing the core motif GGUG. Sequence selectivity resides in a non-canonical interface in RRM2 that is highly related to the SRSF1 pseudoRRM. The atypical Gbp2/Hrb1 C-terminal RRM domains (RRM3) do not interact with RNA/DNA, likely because of their novel N-terminal extensions that block the canonical RNA binding interface. Instead, we discovered that RRM3 is crucial for interaction with the THO/TREX complex and identified key residues essential for this interaction. Moreover, Gbp2 interacts genetically with Tho2 as the double deletion shows a synthetic phenotype and preventing Gbp2 interaction with the THO/TREX complex partly supresses gene expression defect associated with inactivation of the latter complex. These findings provide structural and functional insights into the contribution of SR-like proteins in the post-transcriptional control of gene expression.
Organizational Affiliation:
Department of Biological Physical Chemistry, Instituto de Química-Física 'Rocasolano', CSIC, Serrano-119, 28006 Madrid, Spain.