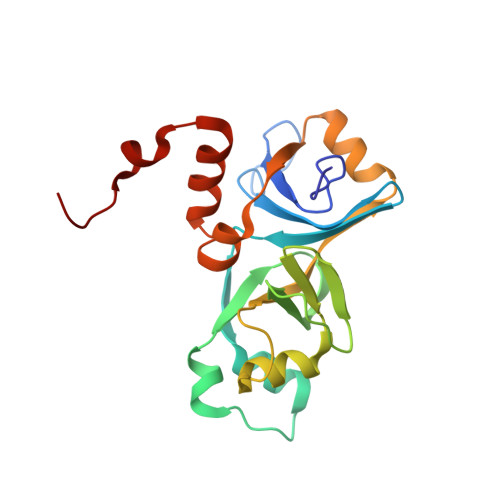
Solution structure and properties of AlgH from Pseudomonas aeruginosa.
Urbauer, J.L., Cowley, A.B., Broussard, H.P., Niedermaier, H.T., Bieber Urbauer, R.J.(2015) Proteins 83: 1137-1150
- PubMed: 25857636
- DOI: https://doi.org/10.1002/prot.24811
- Primary Citation of Related Structures:
2MUI - PubMed Abstract:
In Pseudomonas aeruginosa, the algH gene regulates the cellular concentrations of a number of enzymes and the production of several virulence factors, and is suggested to serve a global regulatory function. The precise mechanism by which the algH gene product, the AlgH protein, functions is unknown. The same is true for AlgH family members from other bacteria. In order to lay the groundwork for understanding the physical underpinnings of AlgH function, we examined the structure and physical properties of AlgH in solution. Under reducing conditions, results of NMR, electrophoretic mobility, and sedimentation equilibrium experiments indicate AlgH is predominantly monomeric and monodisperse in solution. Under nonreducing conditions intra and intermolecular disulfide bonds form, the latter promoting AlgH oligomerization. The high-resolution solution structure of AlgH reveals alpha/beta-sandwich architecture fashioned from ten beta strands and seven alpha helices. Comparison with available structures of orthologues indicates conservation of overall structural topology. The region of the protein most strongly conserved structurally also shows the highest amino acid sequence conservation and, as revealed by hydrogen-deuterium exchange studies, is also the most stable. In this region, evolutionary trace analysis identifies two clusters of amino acid residues with the highest evolutionary importance relative to all other AlgH residues. These frame a partially solvent exposed shallow hydrophobic cleft, perhaps identifying a site for intermolecular interactions. The results establish a physical foundation for understanding the structure and function of AlgH and AlgH family proteins and should be of general importance for further investigations of these and related proteins.
Organizational Affiliation:
The Department of Chemistry, the University of Georgia, Athens, Georgia, 30602-2556.