An Unusual Protein-Protein Interaction through Coupled Unfolding and Binding
Yu, T.K., Shin, S.A., Kim, E.H., Kim, S., Ryu, K.S., Cheong, H., Ahn, H.C., Jon, S., Suh, J.Y.(2014) Angew Chem Int Ed Engl 53: 9784-9787
- PubMed: 24985319
- DOI: https://doi.org/10.1002/anie.201404750
- Primary Citation of Related Structures:
2MNU - PubMed Abstract:
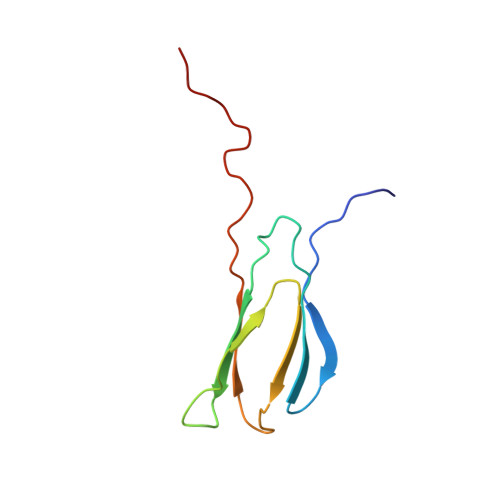
Aptides, a novel class of high-affinity peptides, recognize diverse molecular targets with high affinity and specificity. The solution structure of the aptide APT specifically bound to fibronectin extradomain B (EDB), which represents an unusual protein-protein interaction that involves coupled unfolding and binding, is reported. APT binding is accompanied by unfolding of the C-terminal β strand of EDB, thereby permitting APT to interact with the freshly exposed hydrophobic interior surfaces of EDB. The β-hairpin scaffold of APT drives the interaction by a β-strand displacement mechanism, such that an intramolecular β sheet is replaced by an intermolecular β sheet. The unfolding of EDB perturbs the tight domain association between EDB and FN8 of fibronectin, thus highlighting its potential use as a scaffold that switches between stretched and bent conformations.
Organizational Affiliation:
Biomodulation Major, Department of Agricultural Biotechnology, Seoul National University, 1 Gwanak-ro, Gwanak-gu, Seoul 151-921 (South Korea).