Relationships between IgE/IgG4 epitopes, structure and function in Anisakis simplex Ani s 5, a member of the SXP/RAL-2 protein family
Garcia-Mayoral, M.F., Trevino, M.A., Perez-Pinar, T., Caballero, M.L., Knaute, T., Umpierrez, A., Bruix, M., Rodriguez-Perez, R.(2014) PLoS Negl Trop Dis 8: e2735-e2735
- PubMed: 24603892
- DOI: https://doi.org/10.1371/journal.pntd.0002735
- Primary Citation of Related Structures:
2MAR - PubMed Abstract:
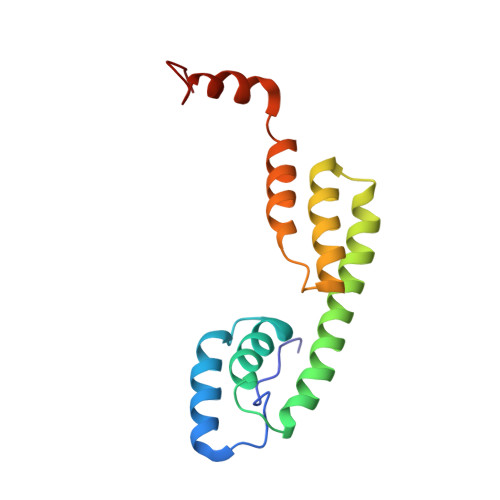
Anisakiasis is a re-emerging global disease caused by consumption of raw or lightly cooked fish contaminated with L3 Anisakis larvae. This zoonotic disease is characterized by severe gastrointestinal and/or allergic symptoms which may misdiagnosed as appendicitis, gastric ulcer or other food allergies. The Anisakis allergen Ani s 5 is a protein belonging to the SXP/RAL-2 family; it is detected exclusively in nematodes. Previous studies showed that SXP/RAL-2 proteins are active antigens; however, their structure and function remain unknown. The aim of this study was to elucidate the three-dimensional structure of Ani s 5 and its main IgE and IgG4 binding regions. The tertiary structure of recombinant Ani s 5 in solution was solved by nuclear magnetic resonance. Mg2+, but not Ca2+, binding was determined by band shift using SDS-PAGE. IgE and IgG4 epitopes were elucidated by microarray immunoassay and SPOTs membranes using sera from nine Anisakis allergic patients. The tertiary structure of Ani s 5 is composed of six alpha helices (H), with a Calmodulin like fold. H3 is a long, central helix that organizes the structure, with H1 and H2 packing at its N-terminus and H4 and H5 packing at its C-terminus. The orientation of H6 is undefined. Regarding epitopes recognized by IgE and IgG4 immunoglobulins, the same eleven peptides derived from Ani s 5 were bound by both IgE and IgG4. Peptides 14 (L40-K59), 26 (A76-A95) and 35 (I103-D122) were recognized by three out of nine sera. This is the first reported 3D structure of an Anisakis allergen. Magnesium ion binding and structural resemblance to Calmodulin, suggest some putative functions for SXP/RAL-2 proteins. Furthermore, the IgE/IgG4 binding regions of Ani s 5 were identified as segments localized on its surface. These data will contribute towards a better understanding of the interactions that occur between immunoglobulins and allergens and, in turn, facilitate the design of novel diagnostic tests and immunotherapeutic strategies.
Organizational Affiliation:
Institute of Physical Chemistry "Rocasolano". CSIC. Madrid, Spain.