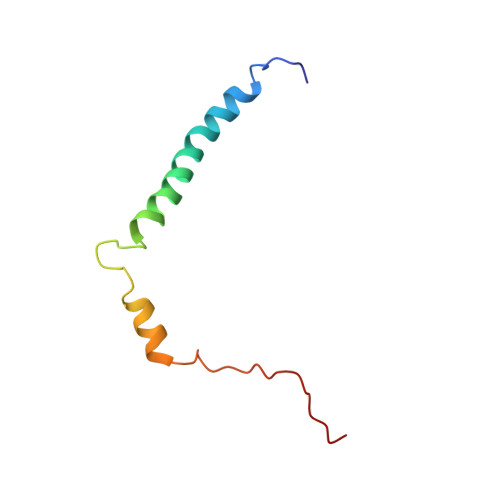
NMR structure of the transmembrane and cytoplasmic domains of human CD4 in micelles.
Wittlich, M., Thiagarajan, P., Koenig, B.W., Hartmann, R., Willbold, D.(2010) Biochim Biophys Acta 1798: 122-127
- PubMed: 19781520
- DOI: https://doi.org/10.1016/j.bbamem.2009.09.010
- Primary Citation of Related Structures:
2KLU - PubMed Abstract:
The human cluster determinant 4 (CD4) is a type I transmembrane glycoprotein involved in T-cell signalling. It is expressed primarily on the surface of T helper cells but also on subsets of memory and regulatory T lymphocytes (CD4(+) cells). It serves as a coreceptor in T-cell receptor recognition of MHC II antigen complexes. Besides its cellular functions, CD4 serves as the main receptor for human immunodeficiency virus type I (HIV-1). During T-cell infection, the CD4 extracellular domain is bound by HIV-1 gp120, the viral surface glycoprotein, which triggers a number of conformational changes ultimately resulting in virion entry of the cell. Subsequently, CD4 is downregulated in infected cells by multiple strategies that involve direct interactions of the HIV-1 proteins VpU and Nef with the cytoplasmic part of CD4. In the present work, we describe the NOE-based solution structure of the transmembrane and cytoplasmic domains of the cystein-free variant of CD4 (CD4mut) in dodecylphosphocholine (DPC) micelles. Furthermore, we have characterized micelle-inserted CD4mut by paramagentic relaxation enhancement (PRE) agents and (1)H-(15)N heteronuclear NOE data. CD4mut features a stable and well-defined transmembrane helix from M372 to V395 buried in the micellar core and a cytoplasmic helix ranging from A404 to L413. Experimental data suggest the amphipathic cytoplasmic helix to be in close contact with the micellar surface. The role of the amphipathic helix and its interaction with the micellar surface is discussed with respect to the biological function of the full-length CD4 protein.
Organizational Affiliation:
Forschungszentrum Jülich, Institut für Strukturbiochemie (ISB-3), 52425 Jülich, Germany.