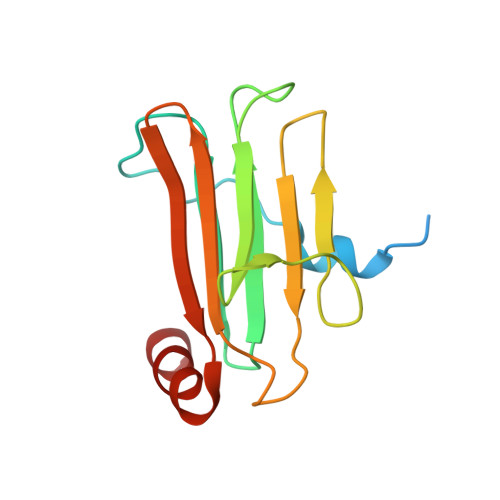
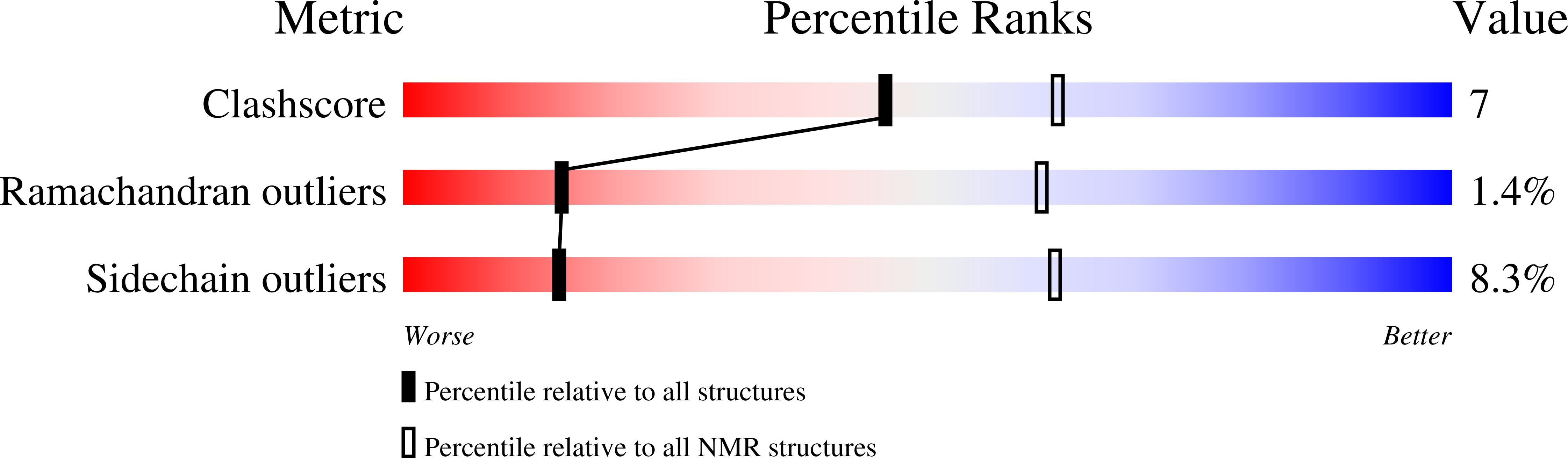Structural insight into the interaction between platelet integrin alphaIIbbeta3 and cytoskeletal protein skelemin.
Deshmukh, L., Tyukhtenko, S., Liu, J., Fox, J.E., Qin, J., Vinogradova, O.(2007) J Biol Chem 282: 32349-32356
- PubMed: 17804417
- DOI: https://doi.org/10.1074/jbc.M704666200
- Primary Citation of Related Structures:
2JTD - PubMed Abstract:
Skelemin is a large cytoskeletal protein critical for cell morphology. Previous studies have suggested that its two-tandem immunoglobulin C2-like repeats (SkIgC4 and SkIgC5) are involved in binding to integrin beta3 cytoplasmic tail (CT), providing a mechanism for skelemin to regulate integrin-mediated signaling and cell spreading. Using NMR spectroscopy, we have studied the molecular details of the skelemin IgC45 interaction with the cytoplasmic face of integrin alphaIIbbeta3. Here, we show that skelemin IgC45 domains form a complex not only with integrin beta3 CT but also, surprisingly, with the integrin alphaIIb CT. Chemical shift mapping experiments demonstrate that both membrane-proximal regions of alphaIIb and beta3 CTs are involved in binding to skelemin. NMR structural determinations, combined with homology modeling, revealed that SkIgC4 and SkIgC5 both exhibited a conserved Ig-fold and both repeats were required for effective binding to and attenuation of alphaIIbbeta3 cytoplasmic complex. These data provide the first molecular insight into how skelemin may interact with integrins and regulate integrin-mediated signaling and cell spreading.
Organizational Affiliation:
Department of Pharmaceutical Sciences, School of Pharmacy, University of Connecticut at Storrs, Storrs, Connecticut 06269, USA.