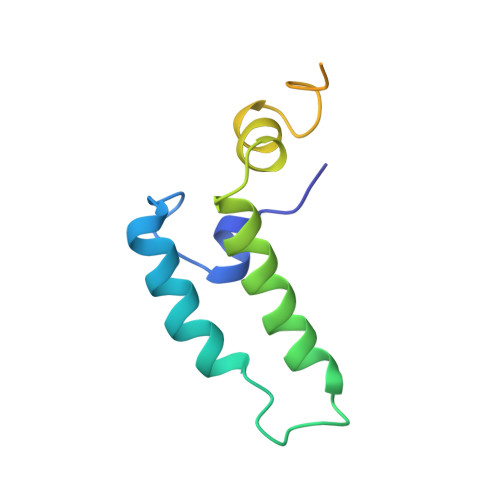
NMR structure of the J-domain and the Gly/Phe-rich region of the Escherichia coli DnaJ chaperone.
Pellecchia, M., Szyperski, T., Wall, D., Georgopoulos, C., Wuthrich, K.(1996) J Mol Biol 260: 236-250
- PubMed: 8764403
- DOI: https://doi.org/10.1006/jmbi.1996.0395
- Primary Citation of Related Structures:
1XBL - PubMed Abstract:
The recombinant N-terminal 107-amino acid polypeptide fragment 2-108 of the DnaJ molecular chaperone of Escherichia coli, which contains the J-domain (residues 2 to 76) and the Gly/Phe-rich region (residues 77 to 108), was uniformly labeled with nitrogen-15 and carbon-13. The complete NMR solution structure of the J-domain was determined with the program DIANA on the basis of 682 nuclear Overhauser enhancement (NOE) upper distance limits and 180 dihedral angle constraints. It contains three well-defined helices comprising residues 6 to 10, 18 to 32 and 41 to 57, and a fourth helix, consisting of residues 61 to 68, which is well defined as a regular secondary structure but for which the location relative to the remainder of the molecule is not precisely determined. The helices II and III form an antiparallel helical coiled-coil. Helix I is approximately parallel to the plane defined by the helices II and III and runs from the carboxy-terminal end of the helix III to the center of helix II. Helix IV is positioned near the carboxy-terminal end of helix III and is on the same side of the coiled coil as helix I, but it is oriented approximately perpendicular to the plane of the helices II and III. This novel alpha-protein topology leads to formation of a hydrophobic core involving side-chains of all four helices. A strong correlation is seen between the extent of sequence-conservation of hydrophobic residues in the family of J-domain homologues, and the structural organization of the hydrophobic core in these proteins. The residues which have key roles for the specificity of the interaction of DnaJ-like proteins with their corresponding Hsp70 counterparts are located on the outer surfaces of the helices II and III, and in the loop connecting these two helices. Measurements of backbone amide proton exchange rates, 15N spin relaxation times and heteronuclear 15N {1H} NOEs provided additional insights into local conformational equilibria and internal rate processes in DnaJ(2-108). In the Gly/Phe-rich region, which is poorly ordered in the NMR solution structure and does not form a globular core, the polypeptide segment 90 to 103 differs from the segments 77 to 89 and 104 to 108 by reduced local flexibility. Considering that this same segment shows sequence conservation with corresponding segments in the Gly/Phe-rich regions of other DnaJ-like proteins, its reduced flexibility may be directly linked to the formation of the ternary DnaJ-DnaK-polypeptide complex.
Organizational Affiliation:
Institut für Molekularbiolgie und Biophysik, Eidgenössische Technische, Zürich, Switzerland.