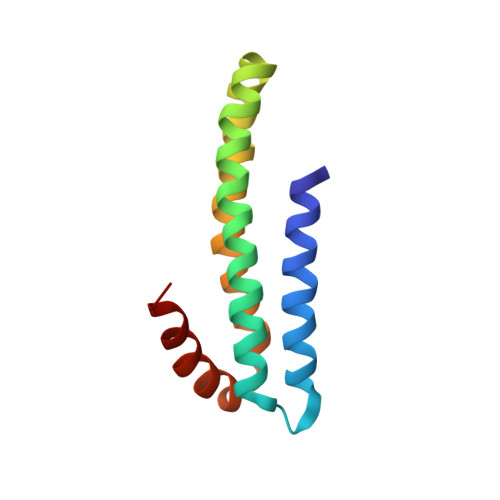
Three-dimensional structure of an independently folded extracellular domain of human amyloid-beta precursor protein.
Dulubova, I., Ho, A., Huryeva, I., Sudhof, T.C., Rizo, J.(2004) Biochemistry 43: 9583-9588
- PubMed: 15274612
- DOI: https://doi.org/10.1021/bi049041o
- Primary Citation of Related Structures:
1TKN - PubMed Abstract:
Cleavage of amyloid-beta precursor protein (APP) by site-specific proteases generates amyloid-beta peptides (Abetas), which are thought to induce Alzheimer's disease. We have identified an independently folded extracellular domain of human APP localized proximal to the Abeta sequence, and determined the three-dimensional structure of this domain by NMR spectroscopy. The domain is composed of four alpha-helices, three of which form a tight antiparallel bundle, and constitutes the C-terminal half of the central extracellular region of APP that has been implicated in the regulation of APP cleavage. Sequence comparisons demonstrate that the domain is highly conserved among all members of the APP family, including invertebrate homologues, suggesting an important role for this region in the biological function of APP. The identification of this domain and the availability of its atomic structure will facilitate analysis of APP function and of the role of the extracellular region in the regulation of APP cleavage.
Organizational Affiliation:
Department of Biochemistry, University of Texas Southwestern Medical Center, 5323 Harry Hines Boulevard, Dallas, Texas 75390, USA. Irina.Dulubova@ UTSouthwestern.edu