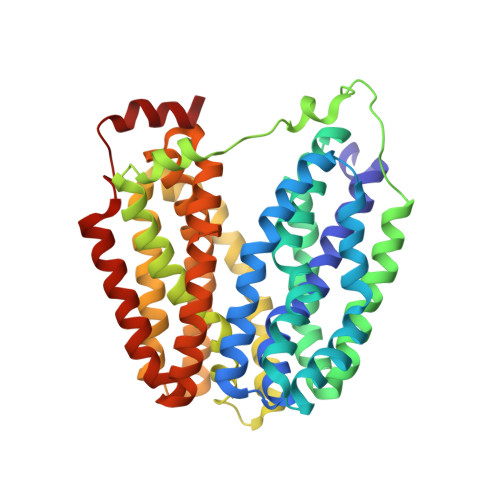
Structure and mechanism of the lactose permease of Escherichia coli
Abramson, J., Smirnova, I., Kasho, V., Verner, G., Kaback, H.R., Iwata, S.(2003) Science 301: 610-615
- PubMed: 12893935
- DOI: https://doi.org/10.1126/science.1088196
- Primary Citation of Related Structures:
1PV6, 1PV7 - PubMed Abstract:
Membrane transport proteins that transduce free energy stored in electrochemical ion gradients into a concentration gradient are a major class of membrane proteins. We report the crystal structure at 3.5 angstroms of the Escherichia coli lactose permease, an intensively studied member of the major facilitator superfamily of transporters. The molecule is composed of N- and C-terminal domains, each with six transmembrane helices, symmetrically positioned within the permease. A large internal hydrophilic cavity open to the cytoplasmic side represents the inward-facing conformation of the transporter. The structure with a bound lactose homolog, beta-D-galactopyranosyl-1-thio-beta-D-galactopyranoside, reveals the sugar-binding site in the cavity, and residues that play major roles in substrate recognition and proton translocation are identified. We propose a possible mechanism for lactose/proton symport (co-transport) consistent with both the structure and a large body of experimental data.
Organizational Affiliation:
Department of Biological Sciences, Imperial College London, London SW7 2AZ, UK.