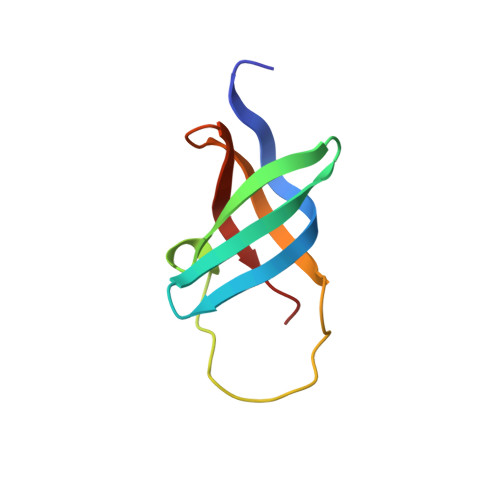
The Solution Structure and DNA-Binding Properties of the Cold-Shock Domain of the Human Y-Box Protein Yb-1.
Kloks, C.P.A.M., Spronk, C.A.E.M., Lasonder, E., Hoffmann, A., Vuister, G.W., Grzesiek, S., Hilbers, C.W.(2002) J Mol Biol 316: 317
- PubMed: 11851341
- DOI: https://doi.org/10.1006/jmbi.2001.5334
- Primary Citation of Related Structures:
1H95 - PubMed Abstract:
The human Y-box protein 1 (YB-1) is a member of the Y-box protein family, a class of proteins involved in transcriptional and translational regulation of a wide range of genes. Here, we report the solution structure of the cold-shock domain (CSD) of YB-1, which is thought to be responsible for nucleic acid binding. It is the first structure solved of a eukaryotic member of the cold-shock protein family and consists of a closed five-stranded anti-parallel beta-barrel capped by a long flexible loop. The structure of CSD is similar to the OB-fold and a comparison with bacterial cold-shock proteins shows that its structural properties are conserved from bacteria to man. Our data suggest the presence of a DNA-binding site consisting of a patch of positively charged and aromatic residues on the surface of the beta-barrel. Further, it is shown that CSD, which has a preference for binding single-stranded pyrimidine-rich sequences, binds weakly and hardly specifically to DNA. Binding affinities reported for intact YB-1 indicate that domains other than the CSD play a role in DNA binding of YB-1.
Organizational Affiliation:
NSR Center for Molecular Structure, Design and Synthesis, Laboratory of Biophysical Chemistry, University of Nijmegen, Toernooiveld 1, 6525 ED Nijmegen, The Netherlands.