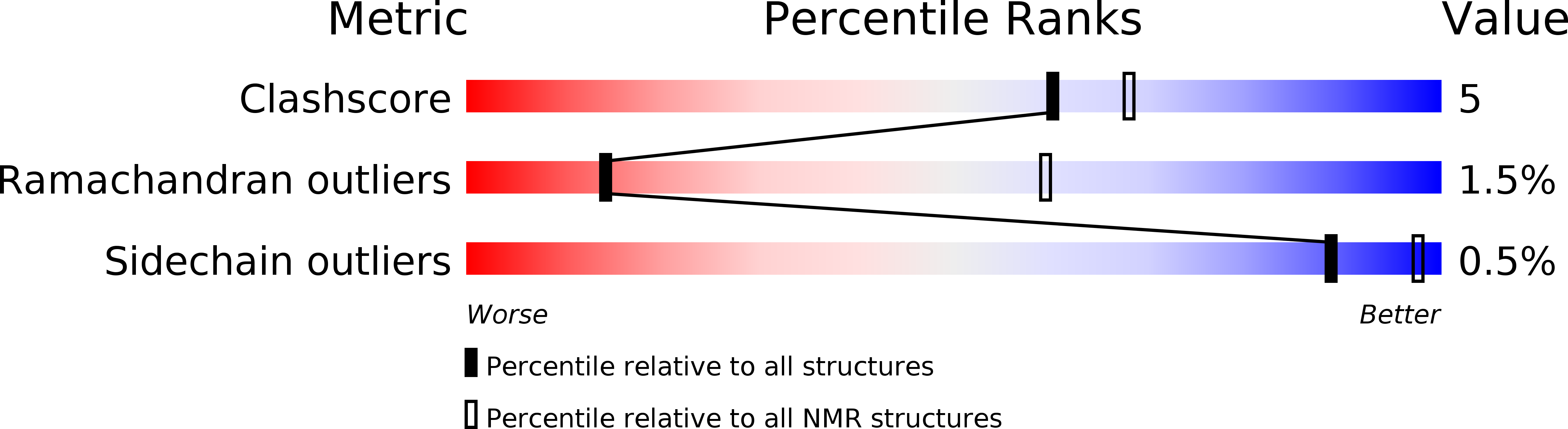
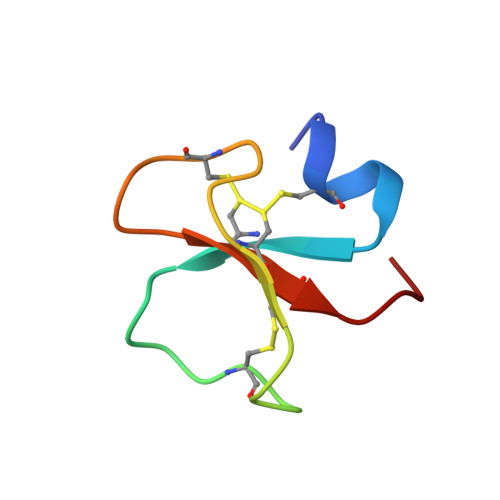
Solution Structure of Crotamine, a Na+ Channel Affecting Toxin from Crotalus Durissus Terrificus Venom
Nicastro, G., Franzoni, L., De Chiara, C., Mancin, A.C.A., Giglio, J.R., Spisni, A.(2003) Eur J Biochem 270: 1969
- PubMed: 12709056
- DOI: https://doi.org/10.1046/j.1432-1033.2003.03563.x
- Primary Citation of Related Structures:
1H5O - PubMed Abstract:
Crotamine is a component of the venom of the snake Crotalus durissus terrificus and it belongs to the myotoxin protein family. It is a 42 amino acid toxin cross-linked by three disulfide bridges and characterized by a mild toxicity (LD50 = 820 micro g per 25 g body weight, i.p. injection) when compared to other members of the same family. Nonetheless, it possesses a wide spectrum of biological functions. In fact, besides being able to specifically modify voltage-sensitive Na+ channel, it has been suggested to exhibit analgesic activity and to be myonecrotic. Here we report its solution structure determined by proton NMR spectroscopy. The secondary structure comprises a short N-terminal alpha-helix and a small antiparallel triple-stranded beta-sheet arranged in an alphabeta1beta2beta3 topology never found among toxins active on ion channels. Interestingly, some scorpion toxins characterized by a biological activity on Na+ channels similar to the one reported for crotamine, exhibit an alpha/beta fold, though with a beta1alphabeta2beta3 topology. In addition, as the antibacterial beta-defensins, crotamine interacts with lipid membranes. A comparison of crotamine with human beta-defensins shows a similar fold and a comparable net positive potential surface. To the best of our knowledge, this is the first report on the structure of a toxin from snake venom active on Na+ channel.
Organizational Affiliation:
Department of Experimental Medicine, Section of Chemistry and Structural Biochemistry, University of Parma, Italy.