Alfa-L-Lna (Alfa-L-Ribo Configured Locked Nucleic Acid) Recognition of DNA: An NMR Spectroscopic Study
Nielsen, K.M.E., Petersen, M., Haakansson, A.E., Wengel, J., Jacobsen, J.P.(2002) Chemistry 8: 3001
- PubMed: 12489231
- DOI: https://doi.org/10.1002/1521-3765(20020703)8:13<3001::AID-CHEM3001>3.0.CO;2-1
- Primary Citation of Related Structures:
1GV6 - PubMed Abstract:
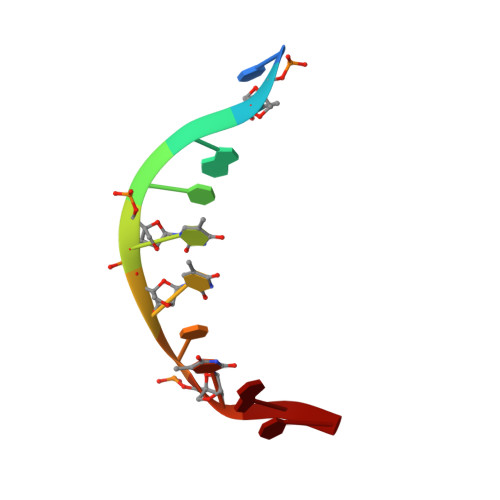
We have used NMR and CD spectroscopy to study and characterise two alpha-L-LNA:DNA duplexes, a nonamer that incorporates three alpha-L-LNA nucleotides and a decamer that incorporates four alpha-L-LNA nucleotides, in which alpha-L-LNA is alpha-L-ribo-configured locked nucleic acid. Both duplexes adopt right-handed helical conformations and form normal Watson-Crick base pairing with all nucleobases in the anti conformation. Deoxyribose conformations were determined from measurements of scalar coupling constants in the sugar rings, and for the decamer duplex, distance information was derived from 1H-1H NOE measurements. In general, the deoxyriboses in both of the alpha-L-LNA:DNA duplexes adopt S-type (B-type structure) sugar puckers, that is the inclusion of the modified alpha-L-LNA nucleotides does not perturb the local native B-like double-stranded DNA (dsDNA) structure. The CD spectra of the duplexes confirm these findings, as these display B-type characteristic features that allow us to characterise the overall duplex type as B-like. The 1H-1H NOE distances which were determined for the decamer duplex were employed in a simulated annealing protocol to generate a model structure for this duplex, thus allowing a more detailed inspection of the impact of the alpha-L-ribo-configured nucleotides. In this structure, it is evident that the malleable DNA backbone rearranges in the vicinity of the modified nucleotides in order to accommodate them and present their nucleobases in a geometry suitable for Watson-Crick base pairing.
Organizational Affiliation:
Nucleic Acid Center, Department of Chemistry, University of Southern Denmark, Odense University, 5230 Odense M, Denmark.