Structure of the intact transactivation domain of the human papillomavirus E2 protein.
Antson, A.A., Burns, J.E., Moroz, O.V., Scott, D.J., Sanders, C.M., Bronstein, I.B., Dodson, G.G., Wilson, K.S., Maitland, N.J.(2000) Nature 403: 805-809
- PubMed: 10693813
- DOI: https://doi.org/10.1038/35001638
- Primary Citation of Related Structures:
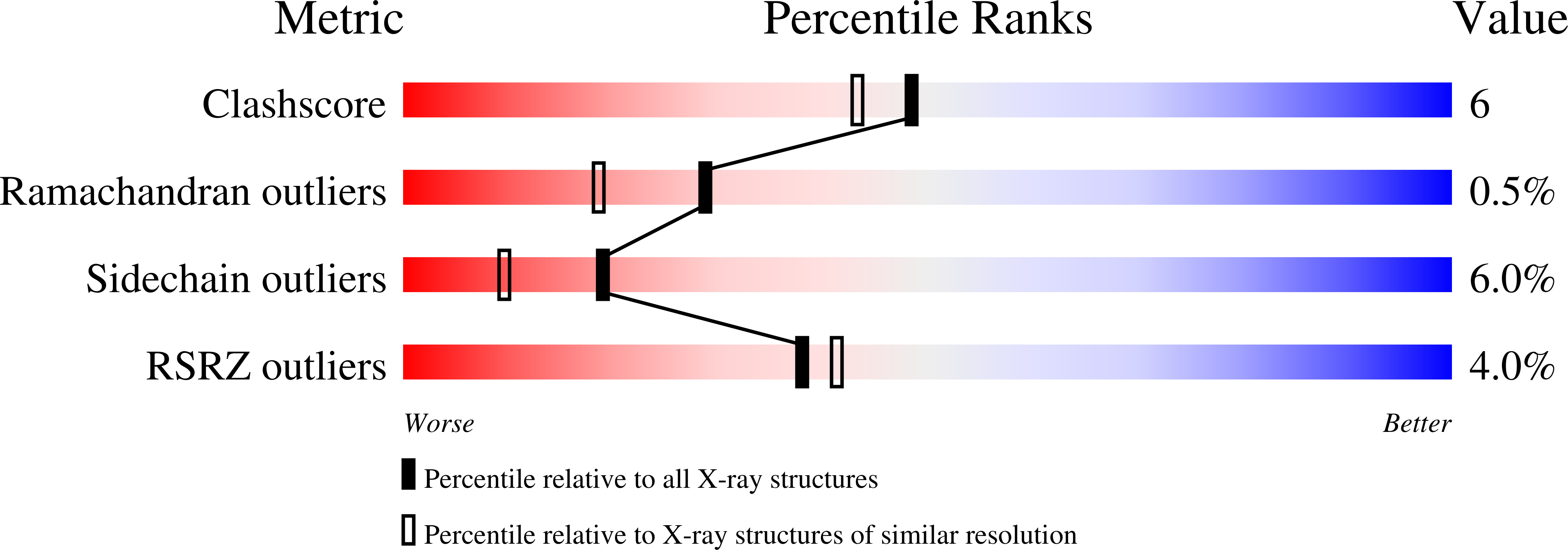
1DTO - PubMed Abstract:
Papillomaviruses cause warts and proliferative lesions in skin and other epithelia. In a minority of papillomavirus types ('high risk, including human papillomaviruses 16, 18, 31, 33, 45 and 56), further transformation of the wart lesions can produce tumours. The papillomavirus E2 protein controls primary transcription and replication of the viral genome. Both activities are governed by a approximately 200 amino-acid amino-terminal module (E2NT) which is connected to a DNA-binding carboxy-terminal module by a flexible linker. Here we describe the crystal structure of the complete E2NT module from human papillomavirus 16. The E2NT module forms a dimer both in the crystal and in solution. Amino acids that are necessary for transactivation are located at the dimer interface, indicating that the dimer structure may be important in the interactions of E2NT with viral and cellular transcription factors. We propose that dimer formation may contribute to the stabilization of DNA loops which may serve to relocate distal DNA-binding transcription factors to the site of human papillomavirus transcription initiation.
Organizational Affiliation:
Department of Chemistry, University of York, UK.