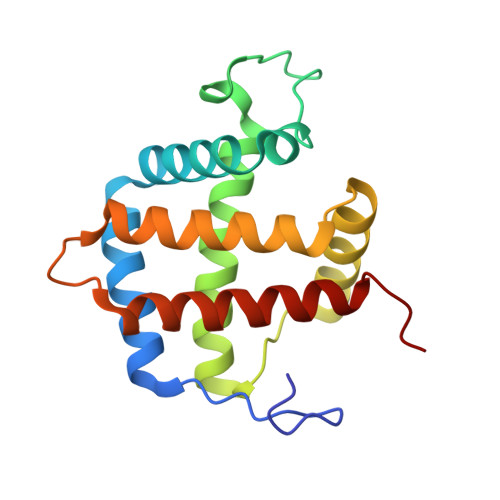
Crystal structure of a nonsymbiotic plant hemoglobin.
Hargrove, M.S., Brucker, E.A., Stec, B., Sarath, G., Arredondo-Peter, R., Klucas, R.V., Olson, J.S., Phillips Jr., G.N.(2000) Structure 8: 1005-1014
- PubMed: 10986467
- DOI: https://doi.org/10.1016/s0969-2126(00)00194-5
- Primary Citation of Related Structures:
1D8U - PubMed Abstract:
Nonsymbiotic hemoglobins (nsHbs) form a new class of plant proteins that is distinct genetically and structurally from leghemoglobins. They are found ubiquitously in plants and are expressed in low concentrations in a variety of tissues including roots and leaves. Their function involves a biochemical response to growth under limited O(2) conditions. The first X-ray crystal structure of a member of this class of proteins, riceHb1, has been determined to 2.4 A resolution using a combination of phasing techniques. The active site of ferric riceHb1 differs significantly from those of traditional hemoglobins and myoglobins. The proximal and distal histidine sidechains coordinate directly to the heme iron, forming a hemichrome with spectral properties similar to those of cytochrome b(5). The crystal structure also shows that riceHb1 is a dimer with a novel interface formed by close contacts between the G helix and the region between the B and C helices of the partner subunit. The bis-histidyl heme coordination found in riceHb1 is unusual for a protein that binds O(2) reversibly. However, the distal His73 is rapidly displaced by ferrous ligands, and the overall O(2) affinity is ultra-high (K(D) approximately 1 nM). Our crystallographic model suggests that ligand binding occurs by an upward and outward movement of the E helix, concomitant dissociation of the distal histidine, possible repacking of the CD corner and folding of the D helix. Although the functional relevance of quaternary structure in nsHbs is unclear, the role of two conserved residues in stabilizing the dimer interface has been identified.
Organizational Affiliation:
Department of Biochemistry, Biophysics & Molecular Biology, Iowa State University, Ames 50011, USA. msh@iastate.edu