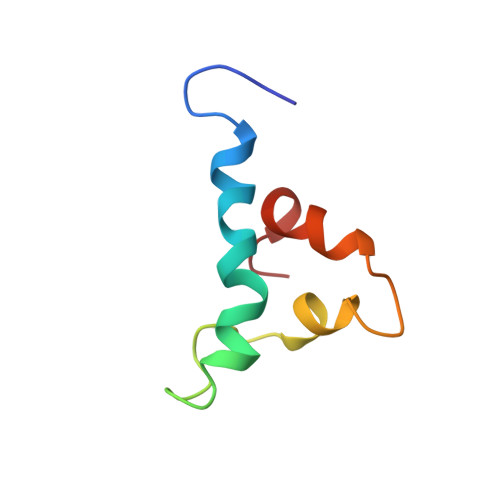
RhoGDI phosphorylation by PKC promotes its interaction with death receptor p75 NTR to gate axon growth and neuron survival.
Ramanujan, A., Li, Z., Ma, Y., Lin, Z., Ibanez, C.F.(2024) EMBO Rep 25: 1490-1512
- PubMed: 38253689
- DOI: https://doi.org/10.1038/s44319-024-00064-2
- Primary Citation of Related Structures:
8X8T - PubMed Abstract:
How receptors juggle their interactions with multiple downstream effectors remains poorly understood. Here we show that the outcome of death receptor p75 NTR signaling is determined through competition of effectors for interaction with its intracellular domain, in turn dictated by the nature of the ligand. While NGF induces release of RhoGDI through recruitment of RIP2, thus decreasing RhoA activity in favor of NFkB signaling, MAG induces PKC-mediated phosphorylation of the RhoGDI N-terminus, promoting its interaction with the juxtamembrane domain of p75 NTR , disengaging RIP2, and enhancing RhoA activity in detriment of NF-kB. This results in stunted neurite outgrowth and apoptosis in cerebellar granule neurons. If presented simultaneously, MAG prevails over NGF. The NMR solution structure of the complex between the RhoGDI N-terminus and p75 NTR juxtamembrane domain reveals previously unknown structures of these proteins and clarifies the mechanism of p75 NTR activation. These results show how ligand-directed competition between RIP2 and RhoGDI for p75 NTR engagement determine axon growth and neuron survival. Similar principles are likely at work in other receptors engaging multiple effectors and signaling pathways.
Organizational Affiliation:
Department of Physiology and Life Sciences Institute, National University of Singapore, 117456, Singapore, Singapore.