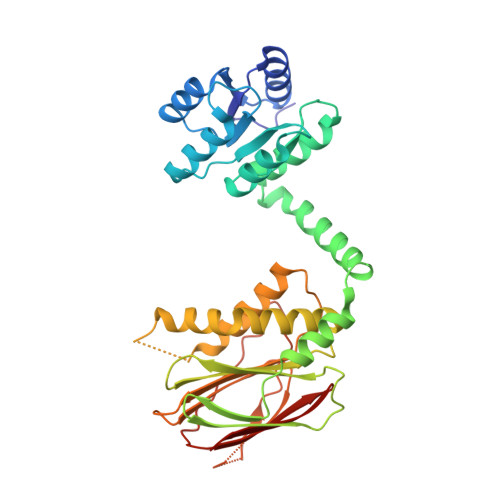
IraM remodels the RssB segmented helical linker to stabilize sigma s against degradation by ClpXP.
Brugger, C., Srirangam, S., Deaconescu, A.M.(2023) J Biol Chem 300: 105568-105568
- PubMed: 38103640
- DOI: https://doi.org/10.1016/j.jbc.2023.105568
- Primary Citation of Related Structures:
8TWD - PubMed Abstract:
Upon Mg 2+ starvation, a condition often associated with virulence, enterobacteria inhibit the ClpXP-dependent proteolysis of the master transcriptional regulator, σ s , via IraM, a poorly understood antiadaptor that prevents RssB-dependent loading of σ s onto ClpXP. This inhibition results in σ s accumulation and expression of stress resistance genes. Here, we report on the structural analysis of RssB bound to IraM, which reveals that IraM induces two folding transitions within RssB, amplified via a segmented helical linker. These conformational changes result in an open, yet inhibited RssB structure in which IraM associates with both the C-terminal and N-terminal domains of RssB and prevents binding of σ s to the 4-5-5 face of the N-terminal receiver domain. This work highlights the remarkable structural plasticity of RssB and reveals how a stress-specific RssB antagonist modulates a core stress response pathway that could be leveraged to control biofilm formation, virulence, and the development of antibiotic resistance.
Organizational Affiliation:
Department of Molecular Biology, Cell Biology and Biochemistry, Brown University, Providence, Rhode Island, USA.