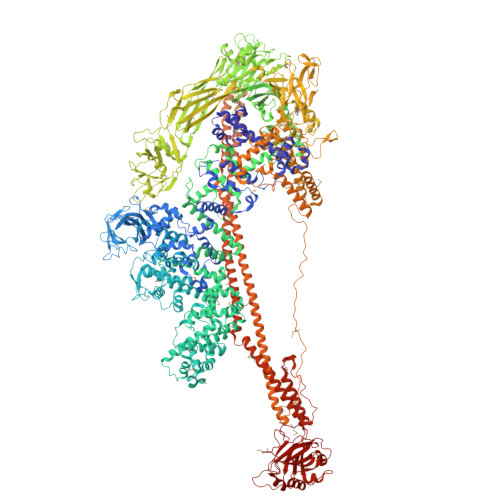
Structures of the Insecticidal Toxin Complex Subunit XptA2 Highlight Roles for Flexible Domains.
Martin, C.L., Chester, D.W., Radka, C.D., Pan, L., Yang, Z., Hart, R.C., Binshtein, E.M., Wang, Z., Nagy, L., DeLucas, L.J., Aller, S.G.(2023) Int J Mol Sci 24
- PubMed: 37686027
- DOI: https://doi.org/10.3390/ijms241713221
- Primary Citation of Related Structures:
8TQE, 8TV0 - PubMed Abstract:
The Toxin Complex (Tc) superfamily consists of toxin translocases that contribute to the targeting, delivery, and cytotoxicity of certain pathogenic Gram-negative bacteria. Membrane receptor targeting is driven by the A-subunit (TcA), which comprises IgG-like receptor binding domains (RBDs) at the surface. To better understand XptA2, an insect specific TcA secreted by the symbiont X. nematophilus from the intestine of entomopathogenic nematodes, we determined structures by X-ray crystallography and cryo-EM. Contrary to a previous report, XptA2 is pentameric. RBD-B exhibits an indentation from crystal packing that indicates loose association with the shell and a hotspot for possible receptor binding or a trigger for conformational dynamics. A two-fragment XptA2 lacking an intact linker achieved the folded pre-pore state like wild type (wt), revealing no requirement of the linker for protein folding. The linker is disordered in all structures, and we propose it plays a role in dynamics downstream of the initial pre-pore state.
Organizational Affiliation:
Department of Pharmacology & Toxicology, University of Alabama at Birmingham, Birmingham, AL 35205, USA.