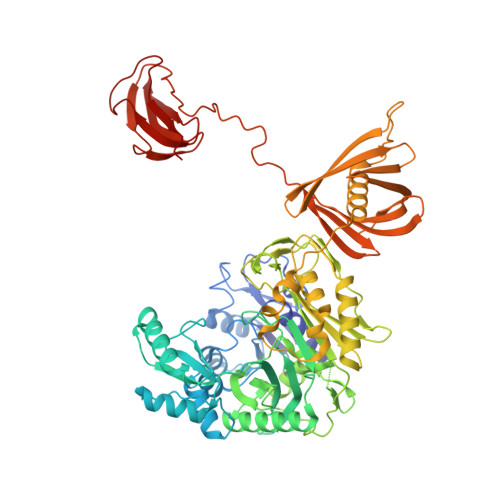
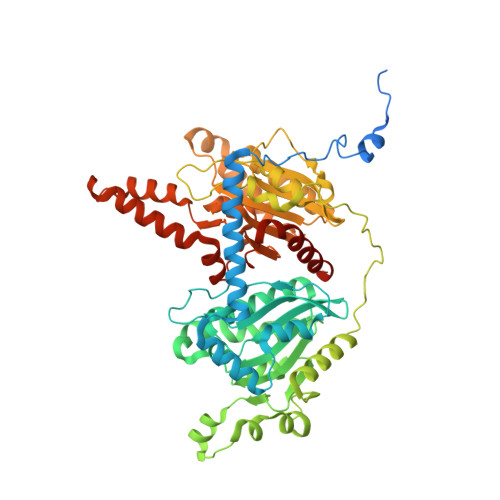
The cryo-EM structure of trypanosome 3-methylcrotonyl-CoA carboxylase provides mechanistic and dynamic insights into its enzymatic function.
Plaza-Pegueroles, A., Aphasizheva, I., Aphasizhev, R., Fernandez-Tornero, C., Ruiz, F.M.(2024) Structure
- PubMed: 38593794
- DOI: https://doi.org/10.1016/j.str.2024.03.010
- Primary Citation of Related Structures:
8RTH - PubMed Abstract:
3-Methylcrotonyl-CoA carboxylase (MCC) catalyzes the two-step, biotin-dependent production of 3-methylglutaconyl-CoA, an essential intermediate in leucine catabolism. Given the critical metabolic role of MCC, deficiencies in this enzyme lead to organic aciduria, while its overexpression is linked to tumor development. MCC is a dodecameric enzyme composed of six copies of each α- and β-subunit. We present the cryo-EM structure of the endogenous MCC holoenzyme from Trypanosoma brucei in a non-filamentous state at 2.4 Å resolution. Biotin is covalently bound to the biotin carboxyl carrier protein domain of α-subunits and positioned in a non-canonical pocket near the active site of neighboring β-subunit dimers. Moreover, flexibility of key residues at α-subunit interfaces and loops enables pivoting of α-subunit trimers to partly reduce the distance between α- and β-subunit active sites, required for MCC catalysis. Our results provide a structural framework to understand the enzymatic mechanism of eukaryotic MCCs and to assist drug discovery against trypanosome infections.
Organizational Affiliation:
Centro de Investigaciones Biológicas Margarita Salas (CIB), CSIC, 28040 Madrid, Spain.