Novel beta-galactosidase activity and first crystal structure of Glycoside Hydrolase family 154.
Hameleers, L., Pijning, T., Gray, B.B., Faure, R., Jurak, E.(2023) N Biotechnol 80: 1-11
- PubMed: 38163476
- DOI: https://doi.org/10.1016/j.nbt.2023.12.011
- Primary Citation of Related Structures:
8OI4 - PubMed Abstract:
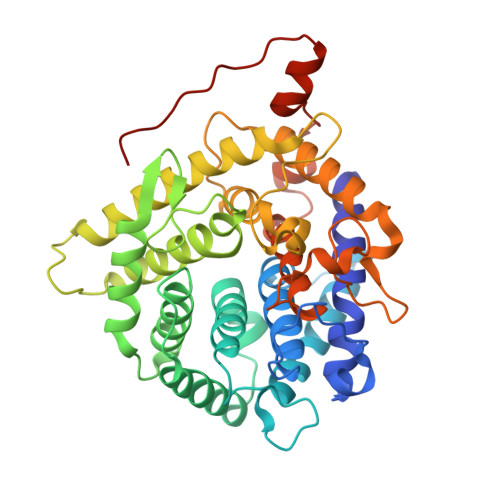
Polysaccharide Utilization Loci (PULs) are physically linked gene clusters conserved in the Gram-negative phylum of Bacteroidota and are valuable sources for Carbohydrate Active enZyme (CAZyme) discovery. This study focuses on BD-β-Gal, an enzyme encoded in a metagenomic PUL and member of the Glycoside Hydrolase family 154 (GH154). BD-β-Gal showed exo-β-galactosidase activity with regiopreference for hydrolyzing β-d-(1,6) glycosidic linkages. Notably, it exhibited a preference for d-glucopyranosyl (d-Glcp) over d-galactopyranosyl (d-Galp) and d-fructofuranosyl (d-Fruf) at the reducing end of the investigated disaccharides. In addition, we determined the high resolution crystal structure of BD-β-Gal, thus providing the first structural characterization of a GH154 enzyme. Surprisingly, this revealed an (α/α) 6 topology, which has not been observed before for β-galactosidases. BD-β-Gal displayed low structural homology with characterized CAZymes, but conservation analysis suggested that the active site was located in a central cavity, with conserved E73, R252, and D253 as putative catalytic residues. Interestingly, BD-β-Gal has a tetrameric structure and a flexible loop from a neighboring protomer may contribute to its reaction specificity. Finally, we showed that the founding member of GH154, BT3677 from Bacteroides thetaiotaomicron, described as β-glucuronidase, displayed exo-β-galactosidase activity like BD-β-Gal but lacked a tetrameric structure.
Organizational Affiliation:
Department of Bioproduct Engineering, Engineering and Technology institute Groningen (ENTEG), University of Groningen, Nijenborgh 4, Groningen 9747 AG, the Netherlands.