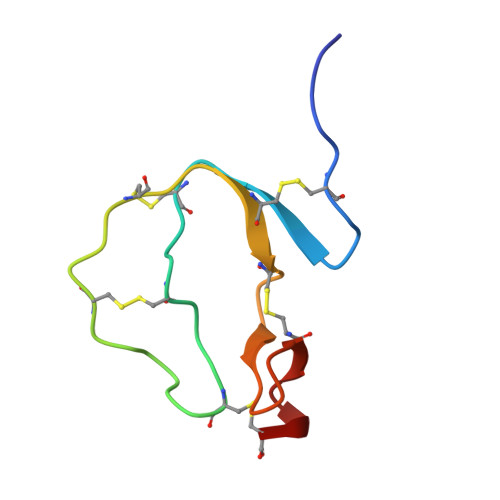
Structural, Functional, and Mutational Studies of a Potent Subtilisin Inhibitor from Budgett's Frog, Lepidobatrachus laevis.
Rami, M., Shafique, M., Sarma, S.P.(2023) Biochemistry 62: 2952-2969
- PubMed: 37796763
- DOI: https://doi.org/10.1021/acs.biochem.3c00252
- Primary Citation of Related Structures:
8HWU, 8J3Q - PubMed Abstract:
Subtilases play a significant role in microbial pathogen infections by degrading the host proteins. Subtilisin inhibitors are crucial in fighting against these harmful microorganisms. LL-TIL, from skin secretions of Lepidobatrachus laevis , is a cysteine-rich peptide belonging to the I8 family of inhibitors. Protease inhibitory assays demonstrated that LL-TIL acts as a slow-tight binding inhibitor of subtilisin Carlsberg and proteinase K with inhibition constants of 91 pM and 2.4 nM, respectively. The solution structures of LL-TIL and a mutant peptide reveal that they adopt a typical TIL-type fold with a canonical conformation of a reactive site loop (RSL). The structure of the LL-TIL-subtilisin complex and molecular dynamics (MD) simulations provided an in-depth view of the structural basis of inhibition. NMR relaxation data and molecular dynamics simulations indicated a rigid conformation of RSL, which does not alter significantly upon subtilisin binding. The energy calculation for subtilisin inhibition predicted Ile 31 as the highest contributor to the binding energy, which was confirmed experimentally by site-directed mutagenesis. A chimeric mutant of LL-TIL broadened the inhibitory profile and attenuated subtilisin inhibition by 2 orders of magnitude. These results provide a template to engineer more specific and potent TIL-type subtilisin inhibitors.
Organizational Affiliation:
Molecular Biophysics Unit, Indian Institute of Science, Bangalore, Karnataka 560012, India.