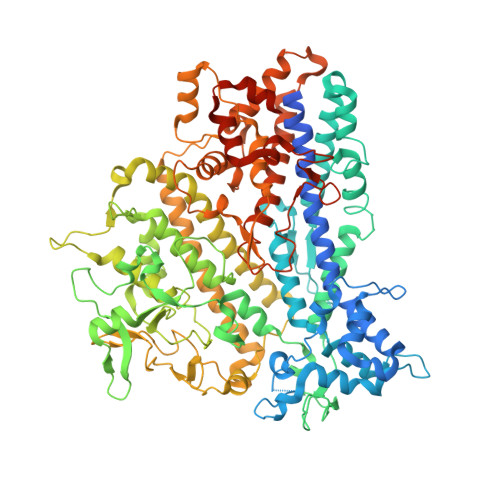
Structural Insights into the Assembly of the African Swine Fever Virus Inner Capsid.
Li, H., Liu, Q., Shao, L., Xiang, Y.(2023) J Virol 97: e0026823-e0026823
- PubMed: 37191520
- DOI: https://doi.org/10.1128/jvi.00268-23
- Primary Citation of Related Structures:
8IGL - PubMed Abstract:
African swine fever virus (ASFV), the cause of a highly contagious hemorrhagic and fatal disease of domestic pigs, has a complex multilayer structure. The inner capsid of ASFV located underneath the inner membrane enwraps the genome-containing nucleoid and is likely the assembly of proteolytic products from the virally encoded polyproteins pp220 and pp62. Here, we report the crystal structure of ASFV p150 △NC , a major middle fragment of the pp220 proteolytic product p150. The structure of ASFV p150 △NC contains mainly helices and has a triangular plate-like shape. The triangular plate is approximately 38 Å in thickness, and the edge of the triangular plate is approximately 90 Å long. The structure of ASFV p150 △NC is not homologous to any of the known viral capsid proteins. Further analysis of the cryo-electron microscopy maps of the ASFV and the homologous faustovirus inner capsids revealed that p150 or the p150-like protein of faustovirus assembles to form screwed propeller-shaped hexametric and pentametric capsomeres of the icosahedral inner capsids. Complexes of the C terminus of p150 and other proteolytic products of pp220 likely mediate interactions between the capsomeres. Together, these findings provide new insights into the assembling of ASFV inner capsid and provide a reference for understanding the assembly of the inner capsids of nucleocytoplasmic large DNA viruses (NCLDV). IMPORTANCE African swine fever virus has caused catastrophic destruction to the pork industry worldwide since it was first discovered in Kenya in 1921. The architecture of ASFV is complicated, with two protein shells and two membrane envelopes. Currently, mechanisms involved in the assembly of the ASFV inner core shell are less understood. The structural studies of the ASFV inner capsid protein p150 performed in this research enable the building of a partial model of the icosahedral ASFV inner capsid, which provides a structural basis for understanding the structure and assembly of this complex virion. Furthermore, the structure of ASFV p150 △NC represents a new type of fold for viral capsid assembly, which could be a common fold for the inner capsid assembly of nucleocytoplasmic large DNA viruses (NCLDV) and would facilitate the development of vaccine and antivirus drugs against these complex viruses.
Organizational Affiliation:
Center for Infectious Disease Research, Beijing Frontier Research Center for Biological Structure & Beijing Advanced Innovation Center for Structural Biology, Department of Basic Medical Sciences, School of Medicine, Tsinghua University, Beijing, China.