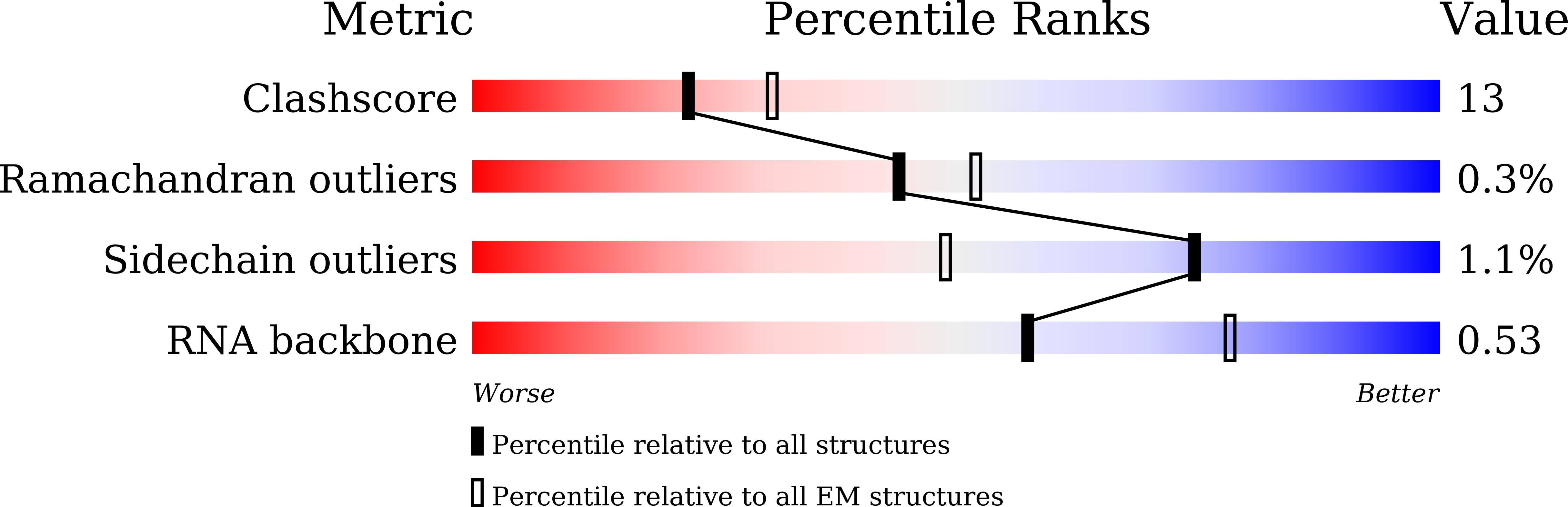
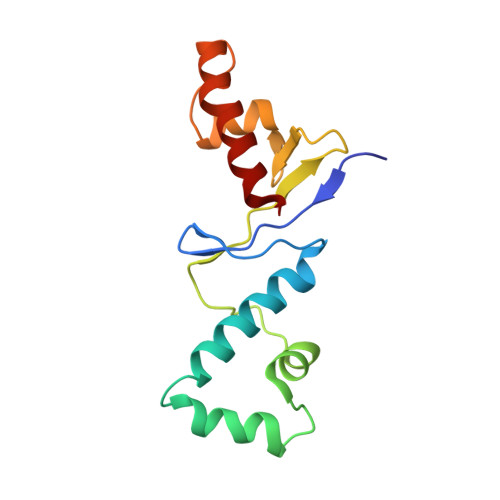
Anti-CRISPR AcrIIC5 is a dsDNA mimic that inhibits type II-C Cas9 effectors by blocking PAM recognition.
Sun, W., Zhao, X., Wang, J., Yang, X., Cheng, Z., Liu, S., Wang, J., Sheng, G., Wang, Y.(2023) Nucleic Acids Res 51: 1984-1995
- PubMed: 36744495
- DOI: https://doi.org/10.1093/nar/gkad052
- Primary Citation of Related Structures:
8HJ4 - PubMed Abstract:
Anti-CRISPR proteins are encoded by phages to inhibit the CRISPR-Cas systems of the hosts. AcrIIC5 inhibits several naturally high-fidelity type II-C Cas9 enzymes, including orthologs from Neisseria meningitidis (Nme1Cas9) and Simonsiella muelleri (SmuCas9). Here, we solve the structure of AcrIIC5 in complex with Nme1Cas9 and sgRNA. We show that AcrIIC5 adopts a novel fold to mimic the size and charge distribution of double-stranded DNA, and uses its negatively charged grooves to bind and occlude the protospacer adjacent motif (PAM) binding site in the target DNA cleft of Cas9. AcrIIC5 is positioned into the crevice between the WED and PI domains of Cas9, and one end of the anti-CRISPR interacts with the phosphate lock loop and a linker between the RuvC and BH domains. We employ biochemical and mutational analyses to build a model for AcrIIC5's mechanism of action, and identify residues on both the anti-CRISPR and Cas9 that are important for their interaction and inhibition. Together, the structure and mechanism of AcrIIC5 reveal convergent evolution among disparate anti-CRISPR proteins that use a DNA-mimic strategy to inhibit diverse CRISPR-Cas surveillance complexes, and provide new insights into a tool for potent inhibition of type II-C Cas9 orthologs.
Organizational Affiliation:
Key Laboratory of RNA Biology, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China.