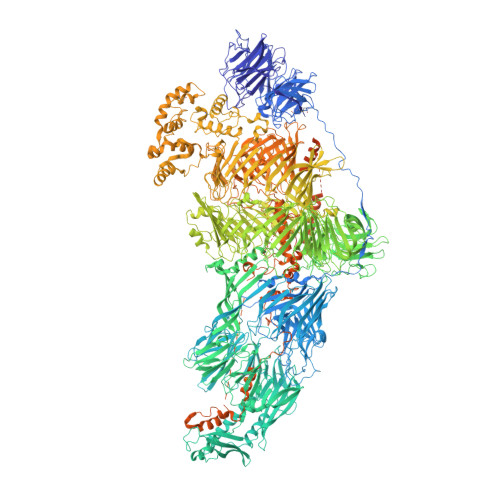
Native structure of mosquito salivary protein uncovers domains relevant to pathogen transmission.
Liu, S., Xia, X., Calvo, E., Zhou, Z.H.(2023) Nat Commun 14: 899-899
- PubMed: 36797290
- DOI: https://doi.org/10.1038/s41467-023-36577-y
- Primary Citation of Related Structures:
8FJP - PubMed Abstract:
Female mosquitoes inject saliva into vertebrate hosts during blood feeding. This process transmits mosquito-borne human pathogens that collectively cause ~1,000,000 deaths/year. Among the most abundant and conserved proteins secreted by female salivary glands is a high-molecular weight protein called salivary gland surface protein 1 (SGS1) that facilitates pathogen transmission, but its mechanism remains elusive. Here, we determine the native structure of SGS1 by the cryoID approach, showing that the 3364 amino-acid protein has a Tc toxin-like Rhs/YD shell, four receptor domains, and a set of C-terminal daisy-chained helices. These helices are partially shielded inside the Rhs/YD shell and poised to transform into predicted transmembrane helices. This transformation, and the numerous receptor domains on the surface of SGS1, are likely key in facilitating sporozoite/arbovirus invasion into the salivary glands and manipulating the host's immune response.
Organizational Affiliation:
Department of Microbiology, Immunology, and Molecular Genetics, University of California, Los Angeles, CA, 90095, USA.