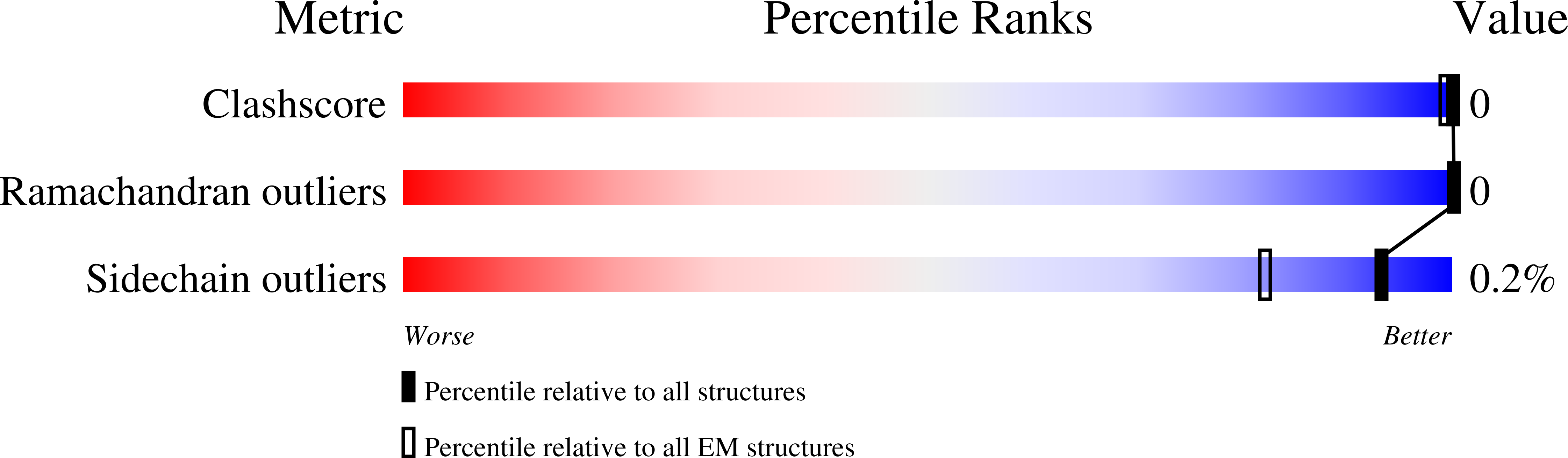Cryo-EM Structure of Gokushovirus Phi EC6098 Reveals a Novel Capsid Architecture for a Single-Scaffolding Protein, Microvirus Assembly System.
Lee, H., Baxter, A.J., Bator, C.M., Fane, B.A., Hafenstein, S.L.(2022) J Virol 96: e0099022-e0099022
- PubMed: 36255280
- DOI: https://doi.org/10.1128/jvi.00990-22
- Primary Citation of Related Structures:
8DES - PubMed Abstract:
Ubiquitous and abundant in ecosystems and microbiomes, gokushoviruses constitute a Microviridae subfamily, distantly related to bacteriophages ΦX174, α3, and G4. A high-resolution cryo-EM structure of gokushovirus ΦEC6098 was determined, and the atomic model was built de novo . Although gokushoviruses lack external scaffolding and spike proteins, which extensively interact with the ΦX174 capsid protein, the core of the ΦEC6098 coat protein (VP1) displayed a similar structure. There are, however, key differences. At each ΦEC6098 icosahedral 3-fold axis, a long insertion loop formed mushroom-like protrusions, which have been noted in lower-resolution gokushovirus structures. Hydrophobic interfaces at the bottom of these protrusions may confer stability to the capsid shell. In ΦX174, the N-terminus of the capsid protein resides directly atop the 3-fold axes of symmetry; however, the ΦEC6098 N-terminus stretched across the inner surface of the capsid shell, reaching nearly to the 5-fold axis of the neighboring pentamer. Thus, this extended N-terminus interconnected pentamers on the inside of the capsid shell, presumably promoting capsid assembly, a function performed by the ΦX174 external scaffolding protein. There were also key differences between the ΦX174-like DNA-binding J proteins and its ΦEC6098 homologue VP8. As seen with the J proteins, C-terminal VP8 residues were bound into a pocket within the major capsid protein; however, its N-terminal residues were disordered, likely due to flexibility. We show that the combined location and interaction of VP8's C-terminus and a portion of VP1's N-terminus are reminiscent of those seen with the ΦX174 and α3 J proteins. IMPORTANCE There is a dramatic structural and morphogenetic divide within the Microviridae . The well-studied ΦX174-like viruses have prominent spikes at their icosahedral vertices, which are absent in gokushoviruses. Instead, gokushovirus major coat proteins form extensive mushroom-like protrusions at the 3-fold axes of symmetry. In addition, gokushoviruses lack an external scaffolding protein, the more critical of the two ΦX174 assembly proteins, but retain an internal scaffolding protein. The ΦEC6098 virion suggests that key external scaffolding functions are likely performed by coat protein domains unique to gokushoviruses. Thus, within one family, different assembly paths have been taken, demonstrating how a two-scaffolding protein system can evolve into a one-scaffolding protein system, or vice versa.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Pennsylvania State Universitygrid.29857.31, University Park, Pennsylvania, USA.