The l,d-Transpeptidase Ldt Ab from Acinetobacter baumannii Is Poorly Inhibited by Carbapenems and Has a Unique Structural Architecture.
Toth, M., Stewart, N.K., Smith, C.A., Lee, M., Vakulenko, S.B.(2022) ACS Infect Dis 8: 1948-1961
- PubMed: 35973205
- DOI: https://doi.org/10.1021/acsinfecdis.2c00321
- Primary Citation of Related Structures:
8DA2 - PubMed Abstract:
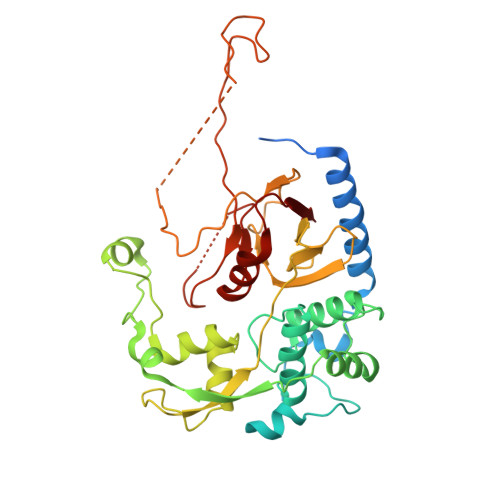
l,d-Transpeptidases (LDTs) are enzymes that catalyze reactions essential for biogenesis of the bacterial cell wall, including formation of 3-3 cross-linked peptidoglycan. Unlike the historically well-known bacterial transpeptidases, the penicillin-binding proteins (PBPs), LDTs are resistant to inhibition by the majority of β-lactam antibiotics, with the exception of carbapenems and penems, allowing bacteria to survive in the presence of these drugs. Here we report characterization of Ldt Ab from the clinically important pathogen, Acinetobacter baumannii . We show that A. baumannii survives inactivation of Ldt Ab alone or in combination with PBP1b or PBP2, while simultaneous inactivation of Ldt Ab and PBP1a is lethal. Minimal inhibitory concentrations (MICs) of all 13 β-lactam antibiotics tested decreased 2- to 8-fold for the Ldt Ab deletion mutant, while further decreases were seen for both double mutants, with the largest, synergistic effect observed for the Ldt Ab + PBP2 deletion mutant. Mass spectrometry experiments showed that Ldt Ab forms complexes in vitro only with carbapenems. However, the acylation rate of these antibiotics is very slow, with the reaction taking longer than four hours to complete. Our X-ray crystallographic studies revealed that Ldt Ab has a unique structural architecture and is the only known LDT to have two different peptidoglycan-binding domains.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of Notre Dame, Notre Dame, Indiana 46556, United States.