Structural and Biochemical Insights into Post-Translational Arginine-to-Ornithine Peptide Modifications by an Atypical Arginase.
Mordhorst, S., Badmann, T., Bosch, N.M., Morinaka, B.I., Rauch, H., Piel, J., Groll, M., Vagstad, A.L.(2023) ACS Chem Biol 18: 528-536
- PubMed: 36791048
- DOI: https://doi.org/10.1021/acschembio.2c00879
- Primary Citation of Related Structures:
8BRP - PubMed Abstract:
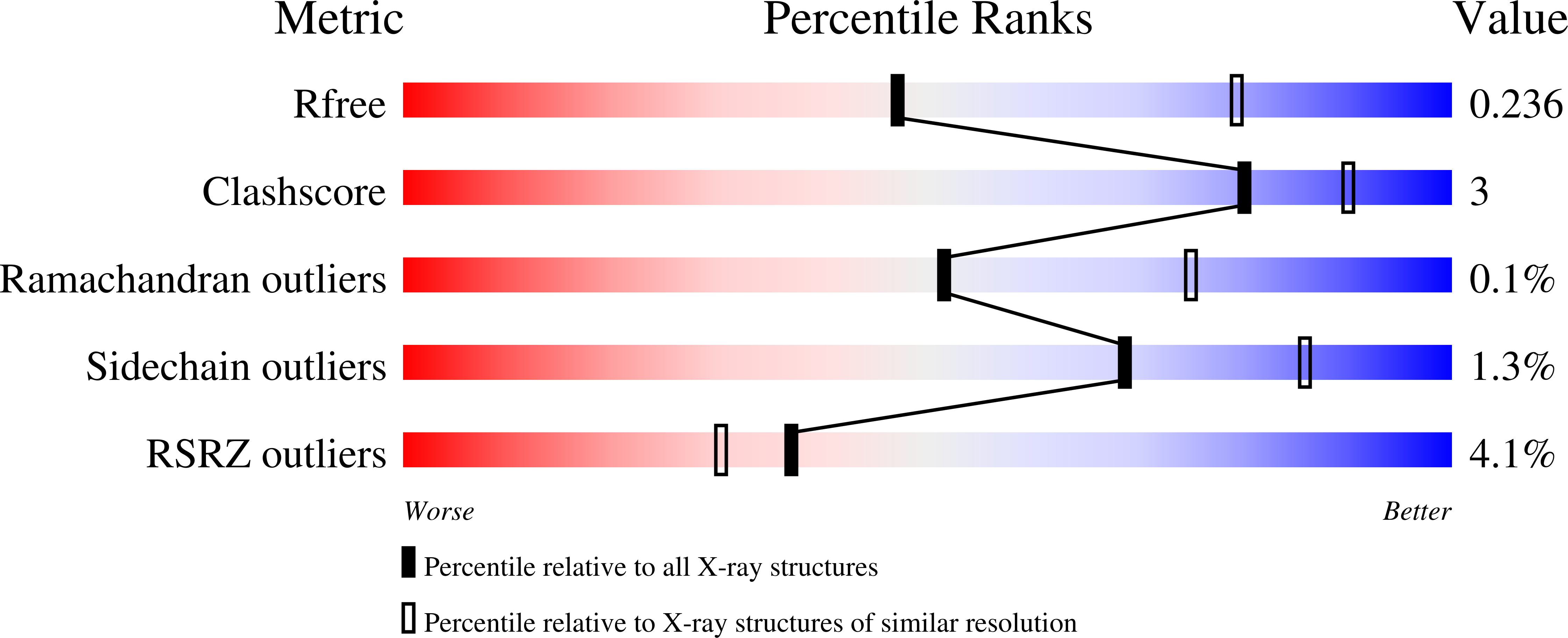
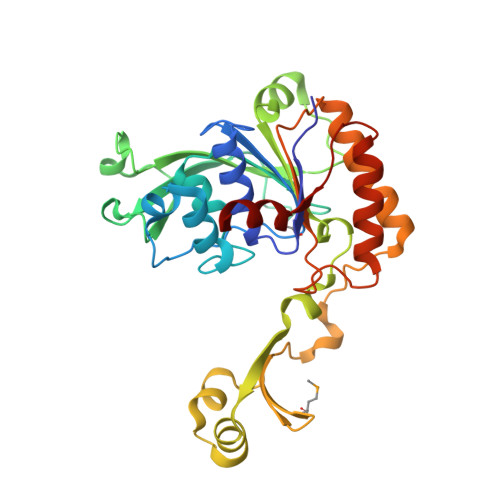
Landornamide A is a ribosomally synthesized and post-translationally modified peptide (RiPP) natural product with antiviral activity. Its biosynthetic gene cluster encodes─among other maturases─the peptide arginase OspR, which converts arginine to ornithine units in an unusual post-translational modification. Peptide arginases are a recently discovered RiPP maturase family with few characterized representatives. They show little sequence similarity to conventional arginases, a well-characterized enzyme family catalyzing the hydrolysis of free arginine to ornithine and urea. Peptide arginases are highly promiscuous and accept a variety of substrate sequences. The molecular basis for binding the large peptide substrate and for the high promiscuity of peptide arginases remains unclear. Here, we report the first crystal structure of a peptide arginase at a resolution of 2.6 Å. The three-dimensional structure reveals common features and differences between conventional arginases and the peptide arginase: the binuclear metal cluster and the active-site environment strongly resemble each other, while the quaternary structures diverge. Kinetic analyses of OspR with various substrates provide new insights into the order of biosynthetic reactions during the post-translational maturation of landornamide A. These results provide the basis for pathway engineering to generate derivatives of landornamide A and for the general application of peptide arginases as biosynthetic tools for peptide engineering.
Organizational Affiliation:
Institute of Microbiology, Eidgenössische Technische Hochschule (ETH) Zürich, Vladimir-Prelog-Weg 4, 8093 Zürich, Switzerland.