Structural Studies of Pif1 Helicases from Thermophilic Bacteria.
Rety, S., Zhang, Y., Fu, W., Wang, S., Chen, W.F., Xi, X.G.(2023) Microorganisms 11
- PubMed: 36838444
- DOI: https://doi.org/10.3390/microorganisms11020479
- Primary Citation of Related Structures:
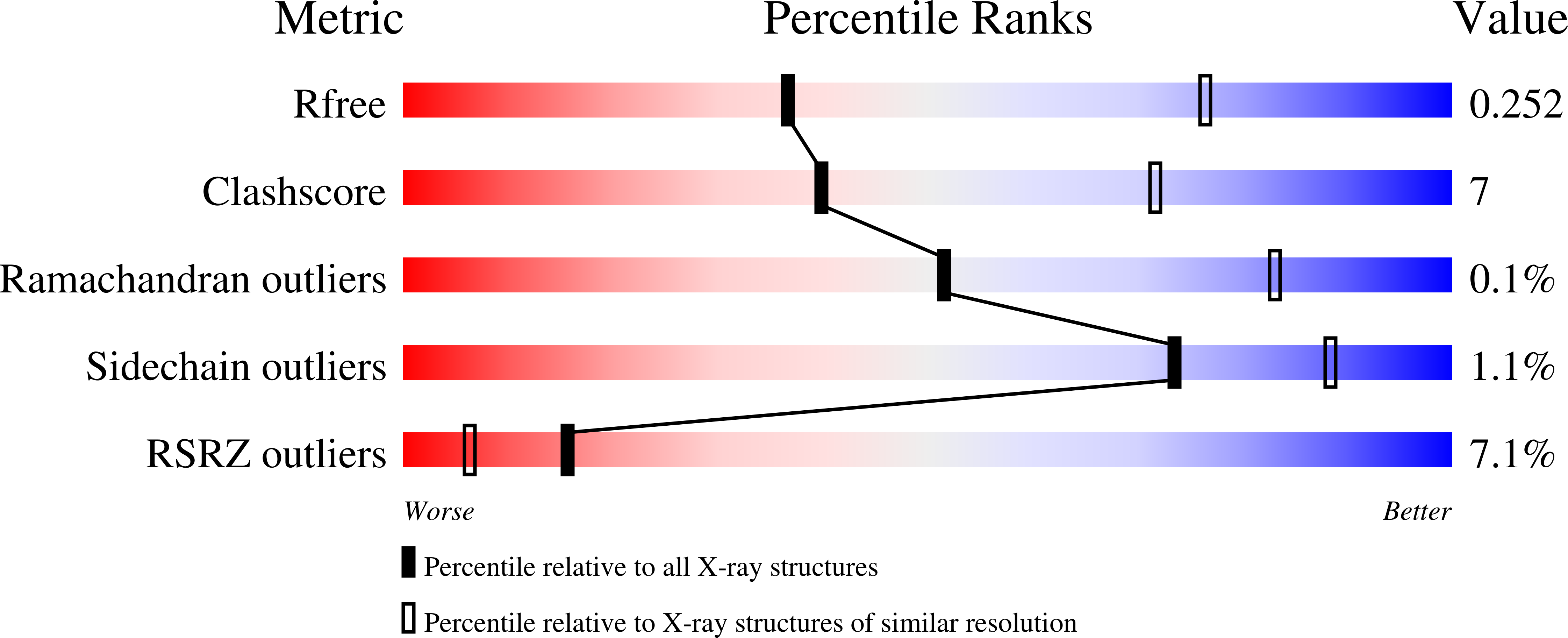
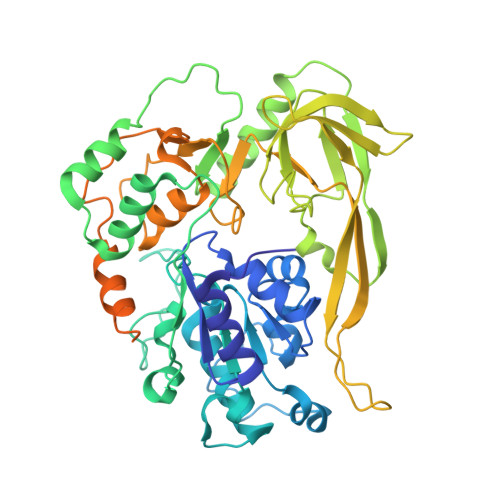
8BNS, 8BNV, 8BNX - PubMed Abstract:
Pif1 proteins are DNA helicases belonging to Superfamily 1, with 5' to 3' directionality. They are conserved from bacteria to human and have been shown to be particularly important in eukaryotes for replication and nuclear and mitochondrial genome stability. However, Pif1 functions in bacteria are less known. While most Pif1 from mesophilic bacteria consist of the helicase core with limited N-terminal and C-terminal extensions, some Pif1 from thermophilic bacteria exhibit a C-terminal WYL domain. We solved the crystal structures of Pif1 helicase cores from thermophilic bacteria Deferribacter desulfuricans and Sulfurihydrogenibium sp. in apo and nucleotide bound form. We show that the N-terminal part is important for ligand binding. The full-length Pif1 helicase was predicted based on the Alphafold algorithm and the nucleic acid binding on the Pif1 helicase core and the WYL domain was modelled based on known crystallographic structures. The model predicts that amino acids in the domains 1A, WYL, and linker between the Helicase core and WYL are important for nucleic acid binding. Therefore, the N-terminal and C-terminal extensions may be necessary to strengthen the binding of nucleic acid on these Pif1 helicases. This may be an adaptation to thermophilic conditions.
Organizational Affiliation:
Laboratoire de Biologie et Modelisation de la Cellule, Ecole Normale Superieure de Lyon, CNRS, UMR 5239, Inserm, U1293, Universite Claude Bernard Lyon 1, 46 allee d'Italie, F-69364 Lyon, France.