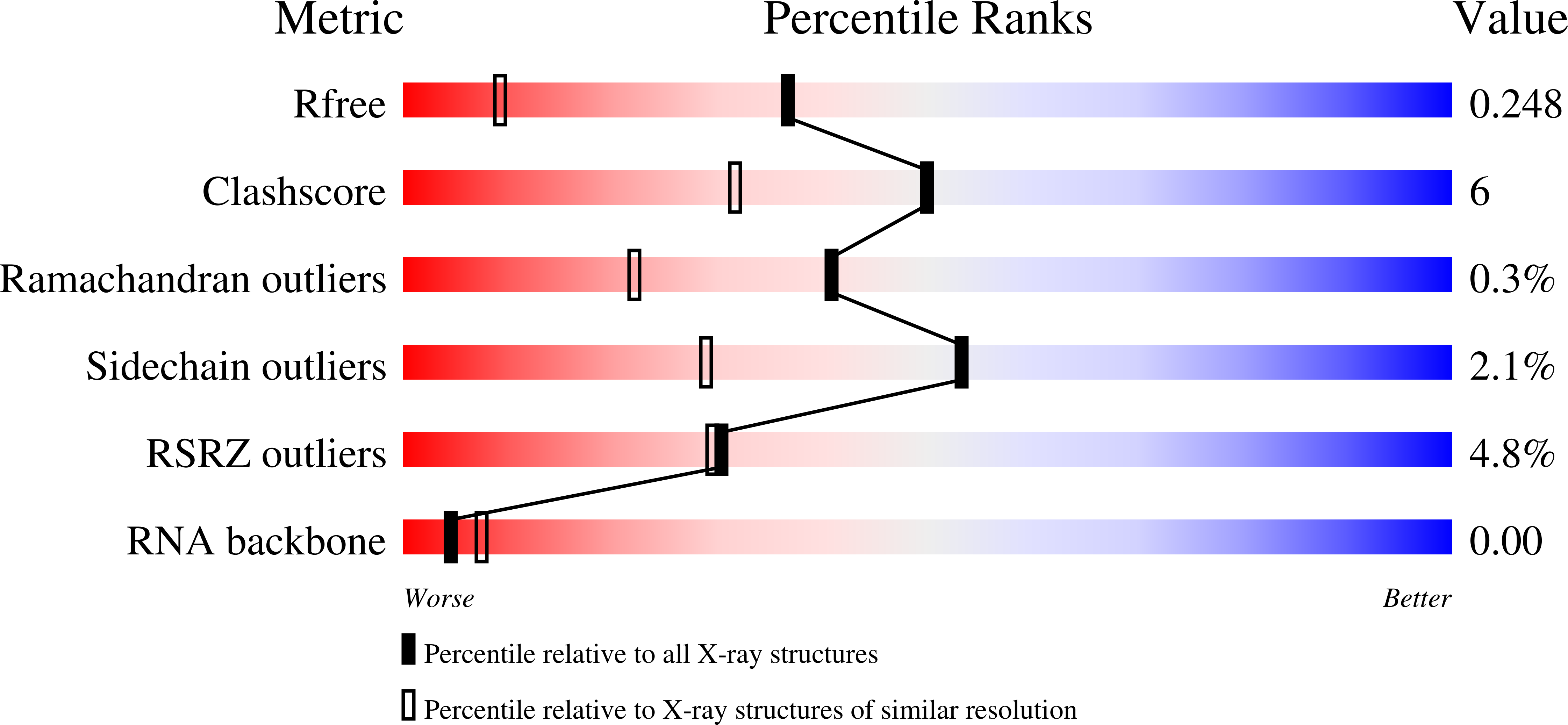
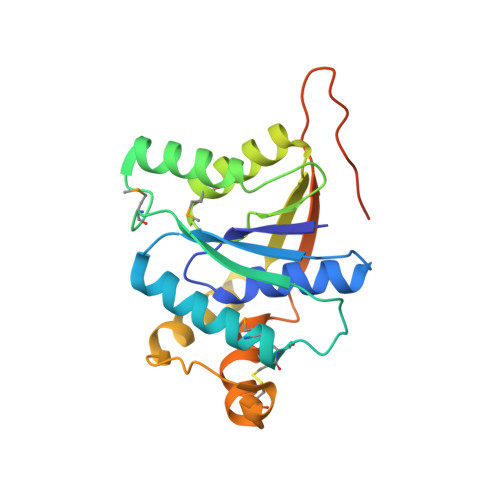
Molecular basis of cyclic tetra-oligoadenylate processing by small standalone CRISPR-Cas ring nucleases.
Molina, R., Garcia-Martin, R., Lopez-Mendez, B., Jensen, A.L.G., Ciges-Tomas, J.R., Marchena-Hurtado, J., Stella, S., Montoya, G.(2022) Nucleic Acids Res 50: 11199-11213
- PubMed: 36271789
- DOI: https://doi.org/10.1093/nar/gkac923
- Primary Citation of Related Structures:
7Z55, 7Z56 - PubMed Abstract:
Standalone ring nucleases are CRISPR ancillary proteins, which downregulate the immune response of Type III CRISPR-Cas systems by cleaving cyclic oligoadenylates (cA) second messengers. Two genes with this function have been found within the Sulfolobus islandicus (Sis) genome. They code for a long polypeptide composed by a CARF domain fused to an HTH domain and a short polypeptide constituted by a CARF domain with a 40 residue C-terminal insertion. Here, we determine the structure of the apo and substrate bound states of the Sis0455 enzyme, revealing an insertion at the C-terminal region of the CARF domain, which plays a key role closing the catalytic site upon substrate binding. Our analysis reveals the key residues of Sis0455 during cleavage and the coupling of the active site closing with their positioning to proceed with cA4 phosphodiester hydrolysis. A time course comparison of cA4 cleavage between the short, Sis0455, and long ring nucleases, Sis0811, shows the slower cleavage kinetics of the former, suggesting that the combination of these two types of enzymes with the same function in a genome could be an evolutionary strategy to regulate the levels of the second messenger in different infection scenarios.
Organizational Affiliation:
Structural Molecular Biology Group, Novo Nordisk Foundation Centre for Protein Research, Faculty of Health and Medical Sciences University of Copenhagen, Blegdamsvej 3-B, 2200 Copenhagen, Denmark.